Testing Extraction of More Pocillopora and Porites Mo'orea Samples
DNA amounts for Pocillopora and Porites Mo’orea samples were really low in the previous extraction, and RNA quality for Porites was poor. We wanted to try bead homogenizing and incubating to see if un-digested proteins/sugars were inhibiting nucleic acid binding on the columns.
Using the Zymo Duet DNA/RNA Extraction Kit
Sample Prep
The starting volume in the tubes (tissue stored in ~300µl DNA/RNA Shield in -20) was a limitation last time when bead homogenizing because it is impossible to get all the liquid out of the tube once the beads are in there. I tried adding 300µl DNA/RNA Shield for 2 of the samples before bead homogenizing to see if this would work better.
| Sample # | Site # | Date Collected | Type | Extraction method |
|---|---|---|---|---|
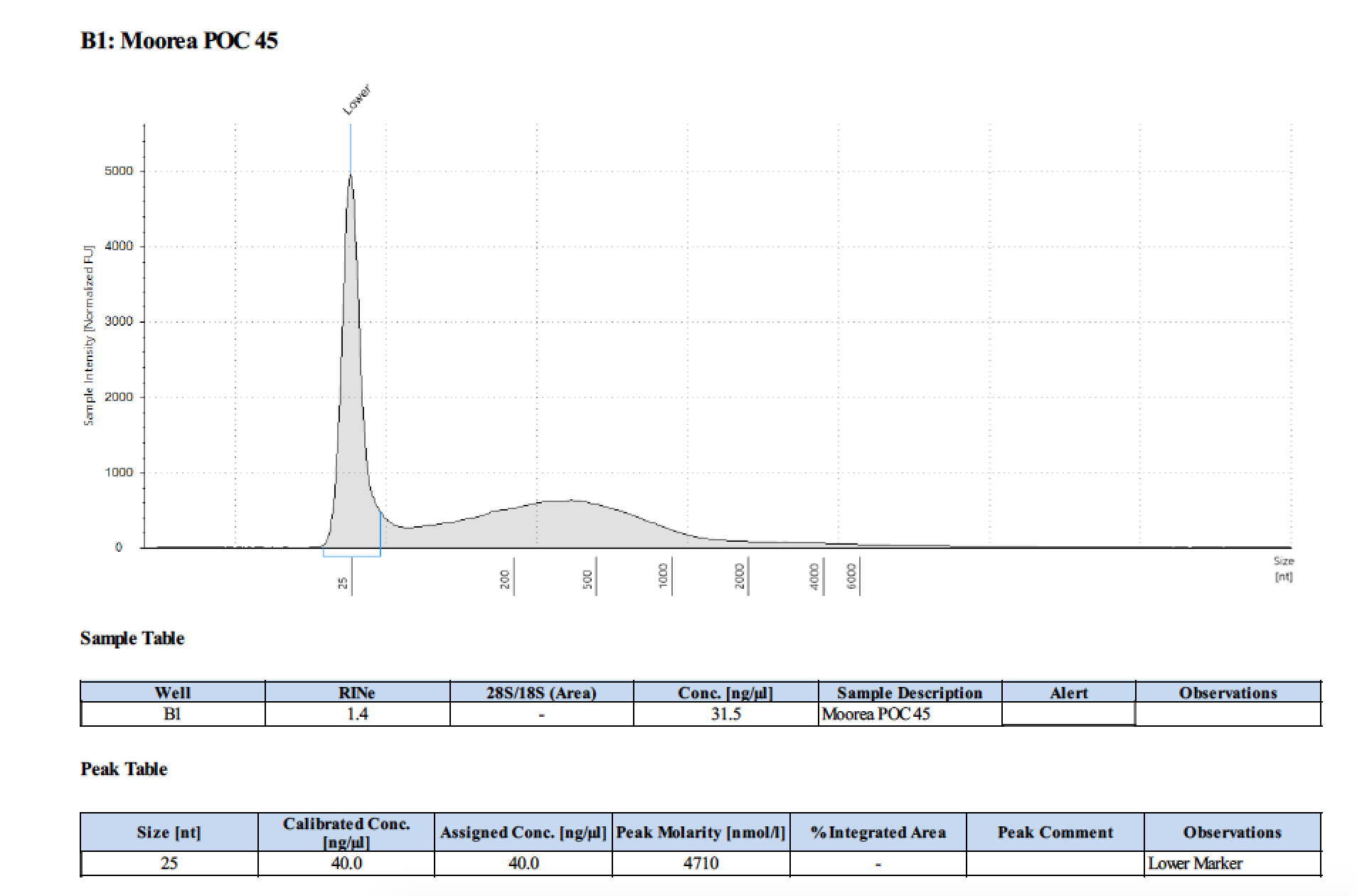
| 45 | 2 | 2018/03/12 | Pocillopora verrucosa | bead homogenize |
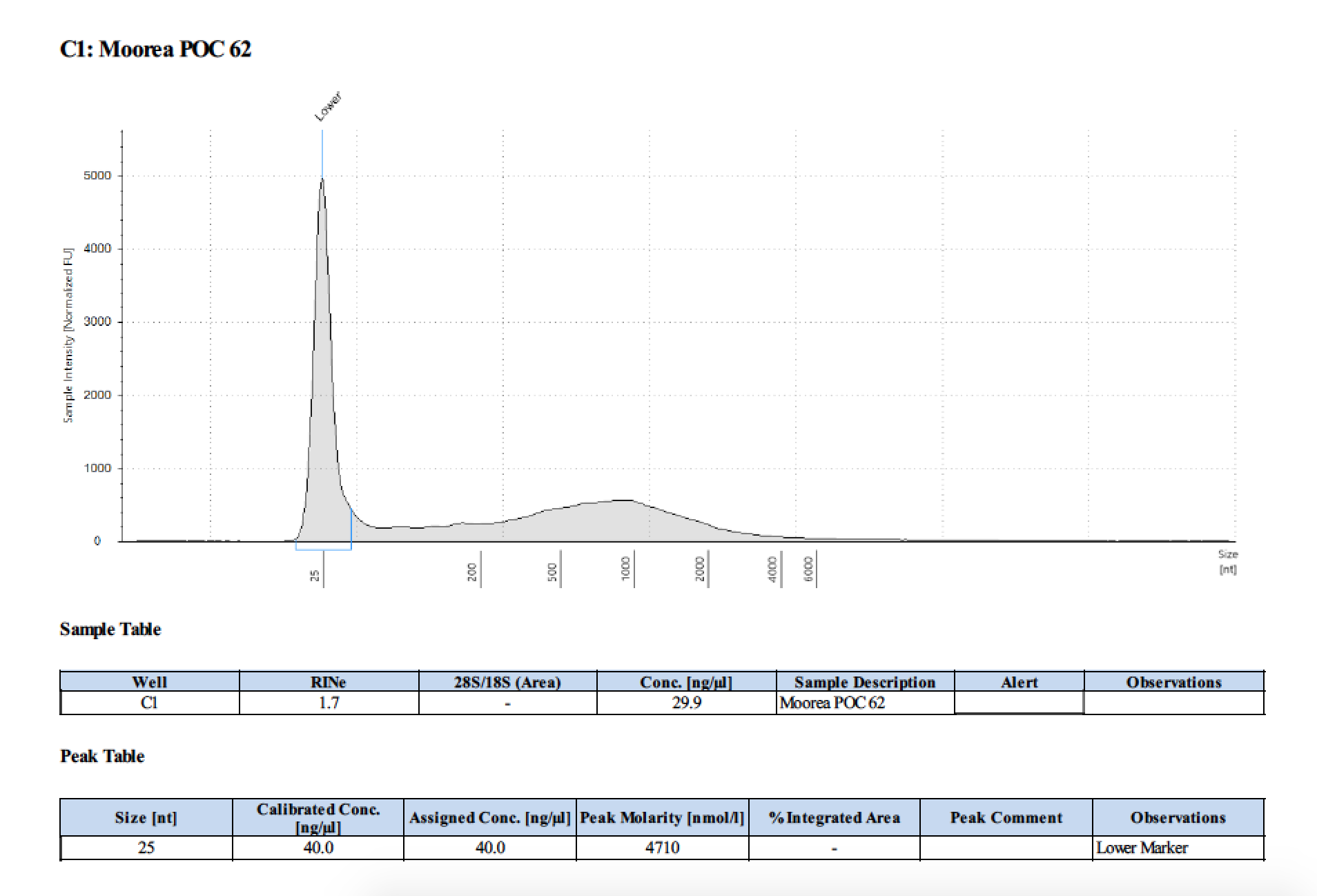
| 62 | 2 | 2018/03/12 | Pocillopora verrucosa | add 300µl then bead homogenize |
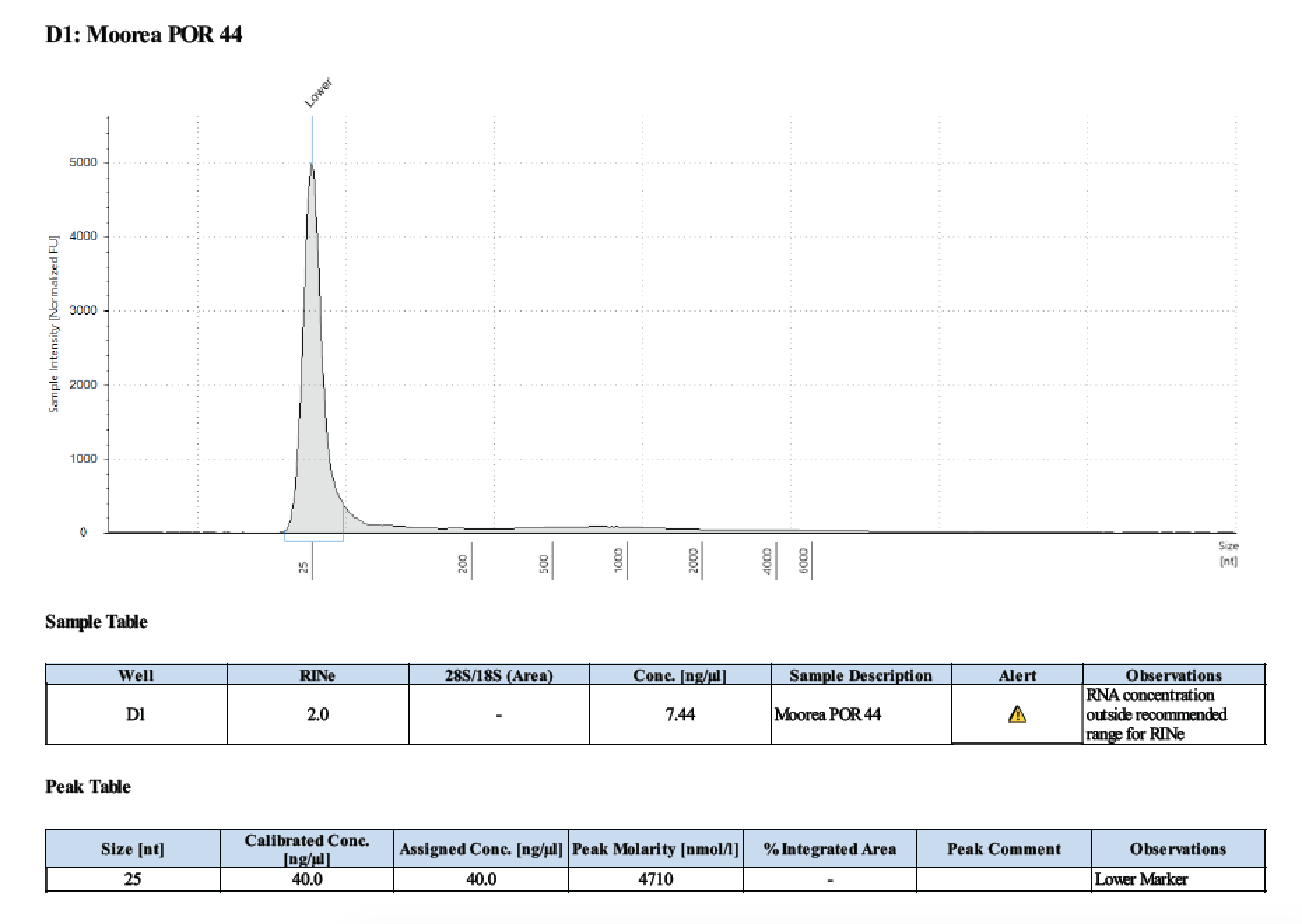
| 44 | 2 | 2018/03/12 | Massive Porites | bead homogenize |
| 63 | 2 | 2018/03/12 | Massive Porites | add 300µl then bead homogenize |
- 300µl of DNA/RNA Shield was added to samples 62 and 63
- Beads from bead tubes were poured into the sample tubes
- All samples were homogenized by vortexing for ~30 seconds
- As much liquid was removed from the tubes as possible and transferred to new 1.5mL tubes
Samples 44 and 63 had mucus mountains making it hard to aspirate the liquid, these are the Porites samples - Appropriate volumes of PK Digestion Buffer and Proteinase K were added based on volumes
| Sample # | Tube Volume | PK Dig Buffer Vol | ProK Vol |
|---|---|---|---|
| 45 | 300µl | 30µl | 15µl |
| 62 | 500µl | 60µl | 30µl |
| 44 | 300µl | 30µl | 15µl |
| 63 | 550µl | 60µl | 30µl |
- Samples were vortexed, spun down, and placed in the Thermomixer at 55 degrees C for 5 hours (max time recommended in the kit) shaking at 1000rpm
DNA Extraction following this mostly
- 10mM Tris HCl and Nuclease-free H20 were put in the incubator at 70 degrees C
- After incubation, all samples were moved to new tubes (some beads had carried over from previous steps)
- ~Equal volumes of DNA/RNA Lysis Buffer were added to each tube;
| Sample # | Vol Lysis Buffer |
|---|---|
| 45 | 345µl |
| 62 | 590µl |
| 44 | 345µl |
| 62 | 640µl |
- Mixed samples by flicking and spinning down
- 600µl of sample was gently added to Yellow DNA spin columns
- Centrifuged columns at 16000 rcf for 30 seconds
-
Flow-through was transferred to new 1.5 (samples 44 and 45) or 5mL (samples 62 and 63) tubes labeled for RNA
- Added 400µl DNA/RNA Prep Buffer gently to the yellow DNA spin columns
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 700µl DNA/RNA Wash Buffer gently to the yellow DNA spin columns
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 400µl DNA/RNA Wash Buffer genetly to the yellow DNA spin columns
- Centrifuged at 16,000 rcf (g) for 2 minutes
- Discarded flow through (Zymo kit waste)
- Transferred yellow columns to new 1.5mL microcentrifuge tubes
- Added 50µl warmed 10mM Tris HCl to each yellow DNA column by dripping slowly directly on the filer
- Incubated at room temp for 5 minutes
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Repeated last three steps for a final elution volume of 100µl
- Labeled tubes, stored at 4 degrees C to quantify and gel the next day
RNA Extraction
- Added equal volume 100% EtOH to the 1.5mL and 5mL tubes labeled for RNA containing the original yellow column flow through
| Sample # | Vol EtOH |
|---|---|
| 45 | 700µl |
| 62 | 1180µl |
| 44 | 700µl |
| 62 | 1200µl |
- Vortexed and spin down to mix
- Added 700µl of that liquid to the green RNA spin columns
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 700µl to the green RNA spin columns (the rest from the 1.5mL RNA tubes)
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 400µl DNA/RNA Wash Buffer gently to each green RNA column
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Made DNase I treatment master mix:
- 75µl DNA Digestion buffer x # of samples
- 5µl DNase I x # of samples
- Added 80µl DNase I treatment master mix directly to the filter of the green RNA columns
- Incubated at room temp for 15 minutes
- Added 400µl DNA/RNA Prep Buffer gently to each column
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 700µl DNA/RNA Wash Buffer gently to the yellow DNA spin columns
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 400µl DNA/RNA Wash Buffer genetly to the yellow DNA spin columns
- Centrifuged at 16,000 rcf (g) for 2 minutes
- Discarded flow through (Zymo kit waste)
- Transferred green columns to new 1.5mL microcentrifuge tubes
- Added 50µl warmed Nuclease free water to each green RNA column by dripping slowly directly on the filer
- Incubated at room temp for 5 minutes
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Repeated last three steps for a final elution volume of 100µl
- Labeled 1.5mL tubes on ice afterwards, and aliquoted 5µl into PCR strip tubes to save for Qubit and Tape Station to avoid freeze-thaw
- Stored all tubes in the -80
Qubit Done on March 21 2019
- Broad Range dsDNA and RNA Qubit protocol
- All samples were read twice
- RNA Qubit was done by Erin Chille
| Sample | DNA Standard 1 (RFU) | DNA Standard 2 (RFU) | DNA 1 (ng/µl) | DNA 2 (ng/µl) | Average DNA | RNA Standard 1 (RFU) | RNA Standard 2 (RFU) | RNA 1 (ng/µl) | RNA 2 (ng/ul) | Average RNA |
|---|---|---|---|---|---|---|---|---|---|---|
| 45 | 205 | 23006 | 29.2 | 28.4 | 28.7 | 406 | 10912 | 17.6 | 17.2 | 17.4 |
| 62 | 205 | 23006 | 30.0 | 29.4 | 29.7 | 406 | 10912 | 35.8 | 36.0 | 35.9 |
| 44 | 205 | 23006 | 5.92 | 5.76 | 5.83 | 406 | 10912 | 44.8 | 45.0 | 44.9 |
| 63 | 205 | 23006 | 22.6 | 22.2 | 22.4 | 406 | 10912 | 53.6 | 53.6 | 53.6 |
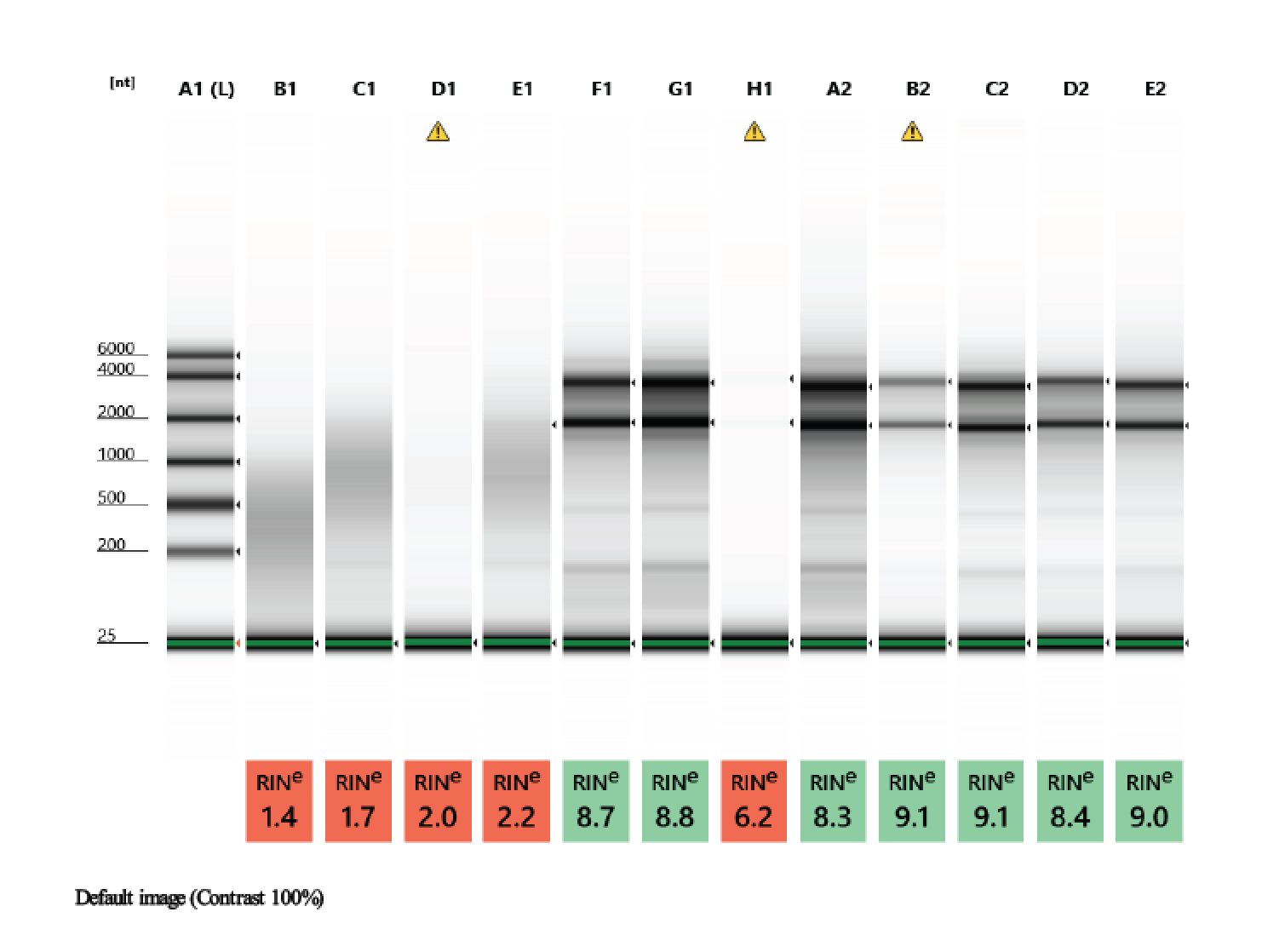
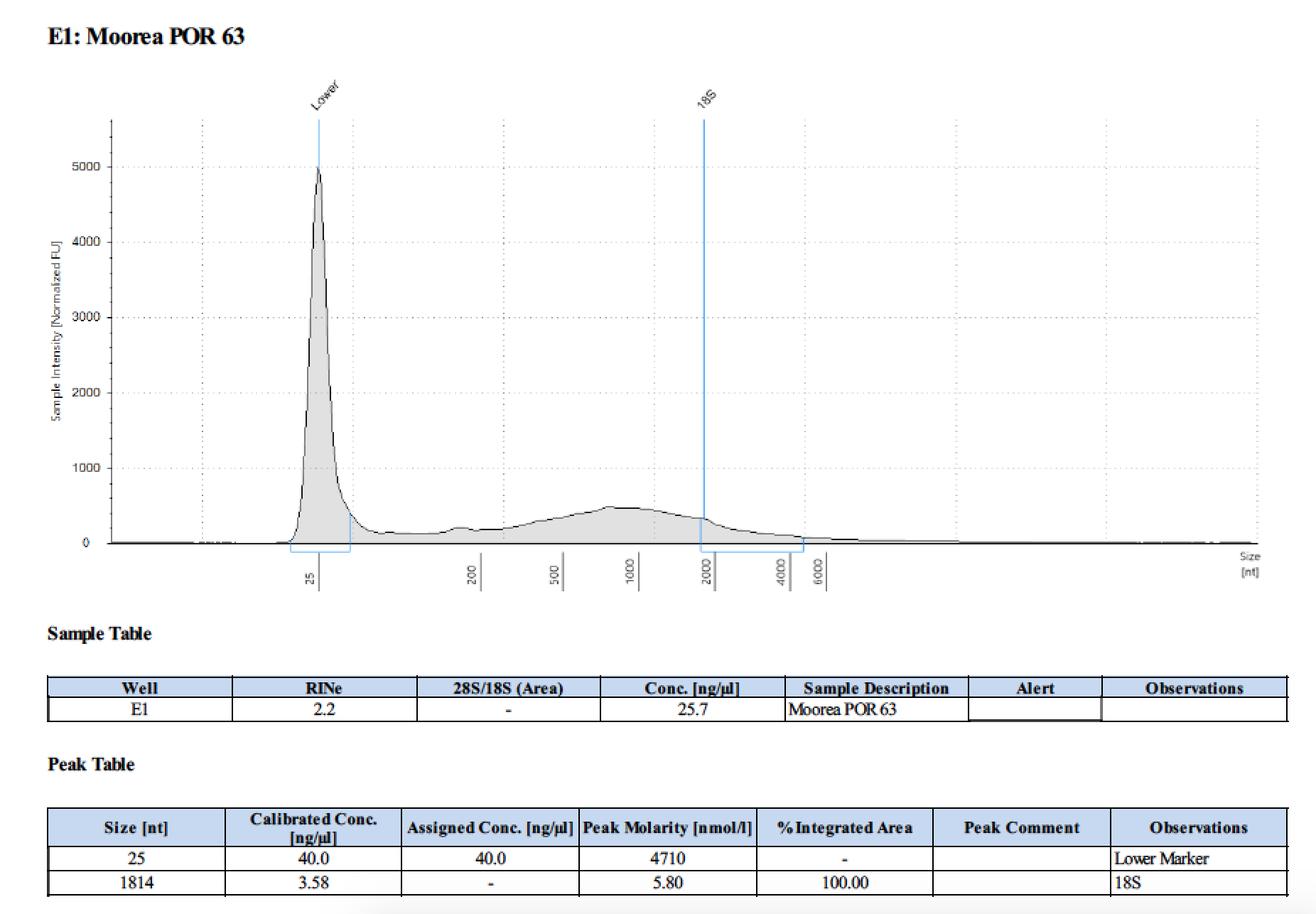
TapeStation
- Followed RNA TapeStation protocol
- TapeStation was done by Erin Chille and include some of her RNA extractions of Montipora larvae
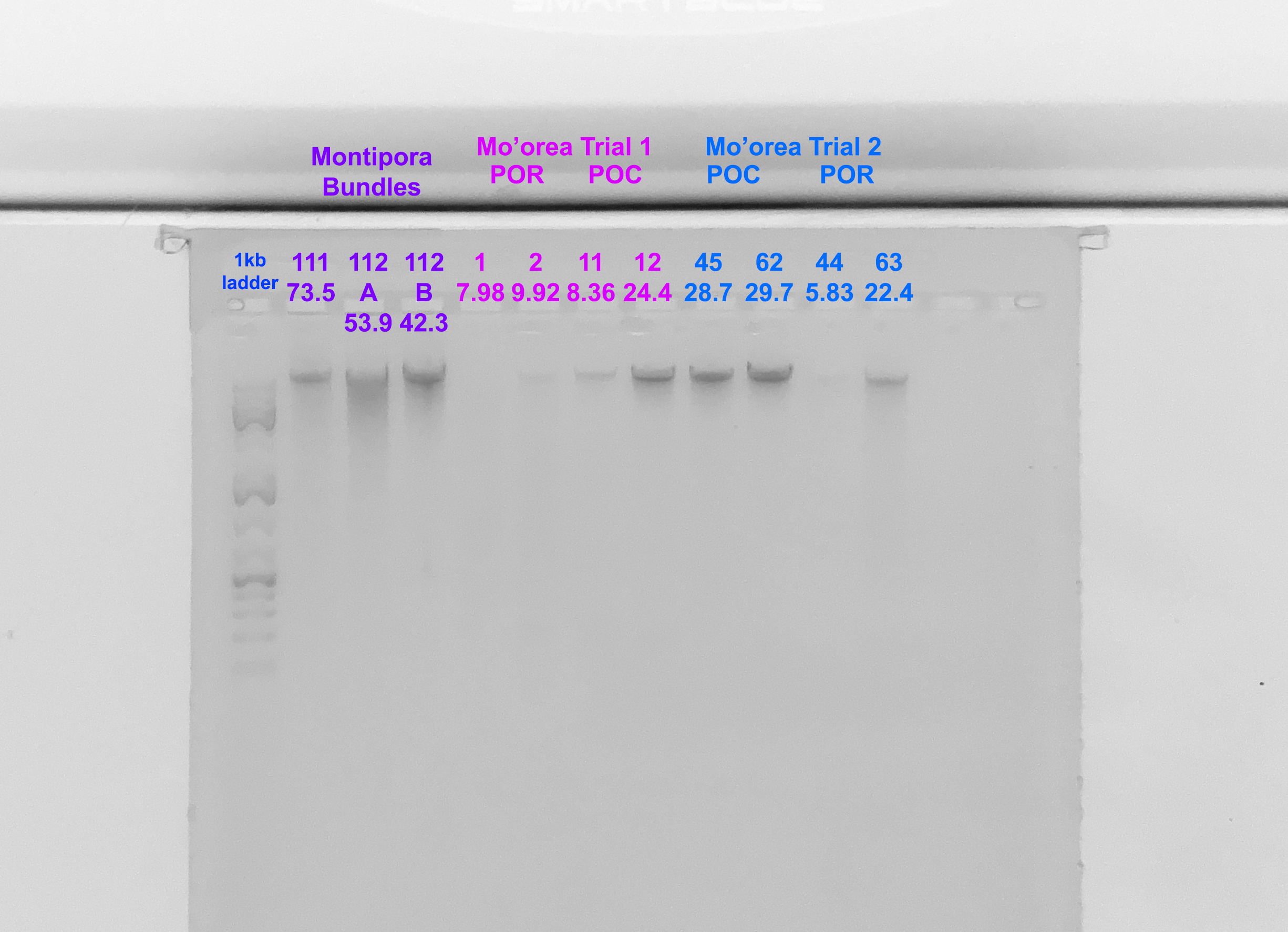
Gel
- A 1.5% agarose gel was ran to check the integrity of the genomic DNA
- Following the PPP Lab protocol
- The first 7 samples are from the previous post, the final 4 are from this extraction
- Below the sample number is the amount of DNA in ng/µl from the Qubit
Written on March 20, 2019