Montipora Eggs and Bundles Troubleshooting and Mo'orea Test DNA/RNA Extraction
Previous DNA/RNA test extractions with the Montipora eggs and bundles from the biomineralization project showed some problems with reliably getting both DNA and RNA from the samples
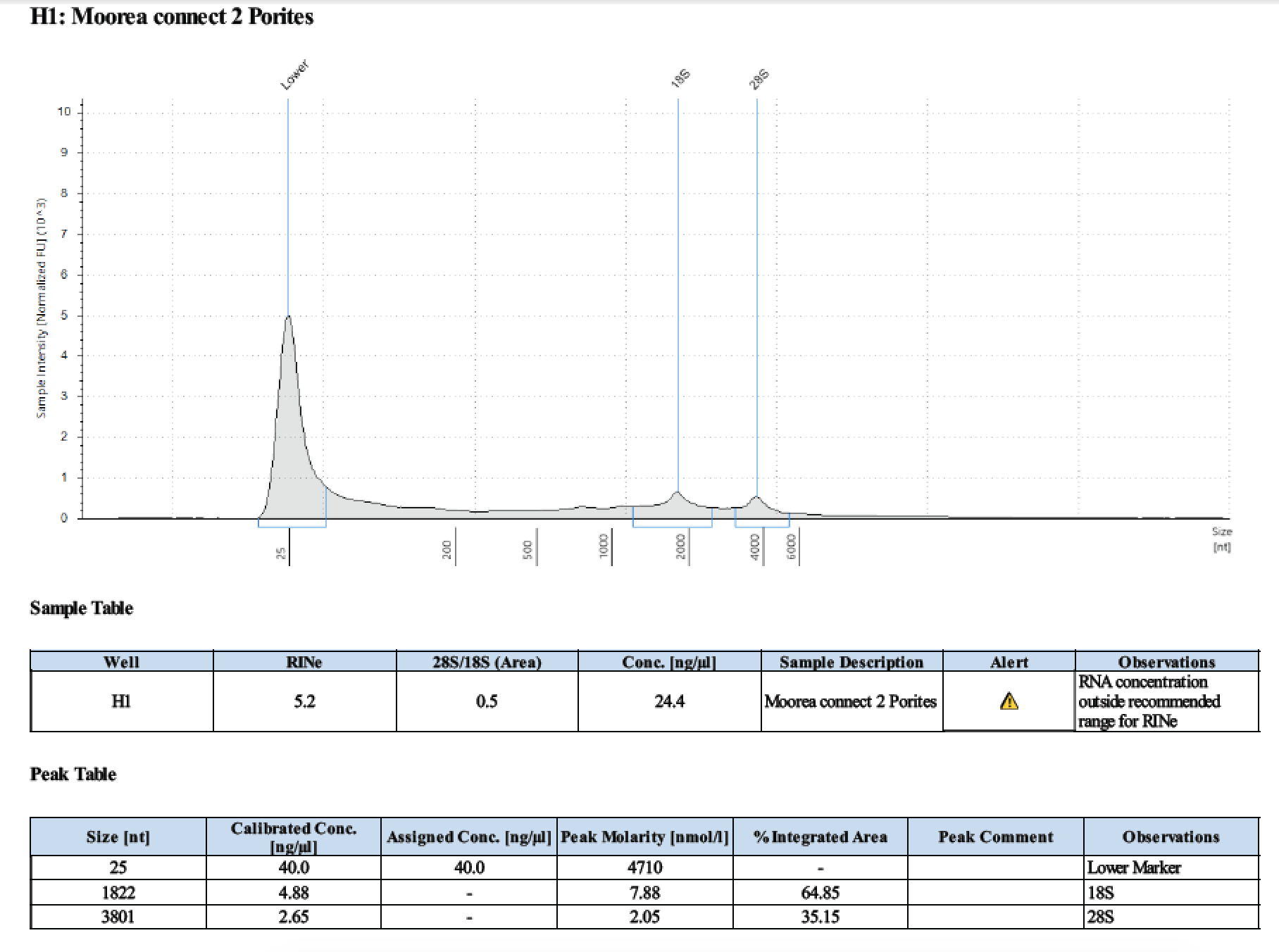
Additionally, Porites and Pocillopora tissue stored in DNA/RNA Shield at -20 from the Putnam-Puritz Mo’orea project needed to have extractions tested on it
Using the Zymo Duet DNA/RNA Extraction Kit
Eggs and Bundles
| Sample # | Date Collected | Type | Extraction method |
|---|---|---|---|
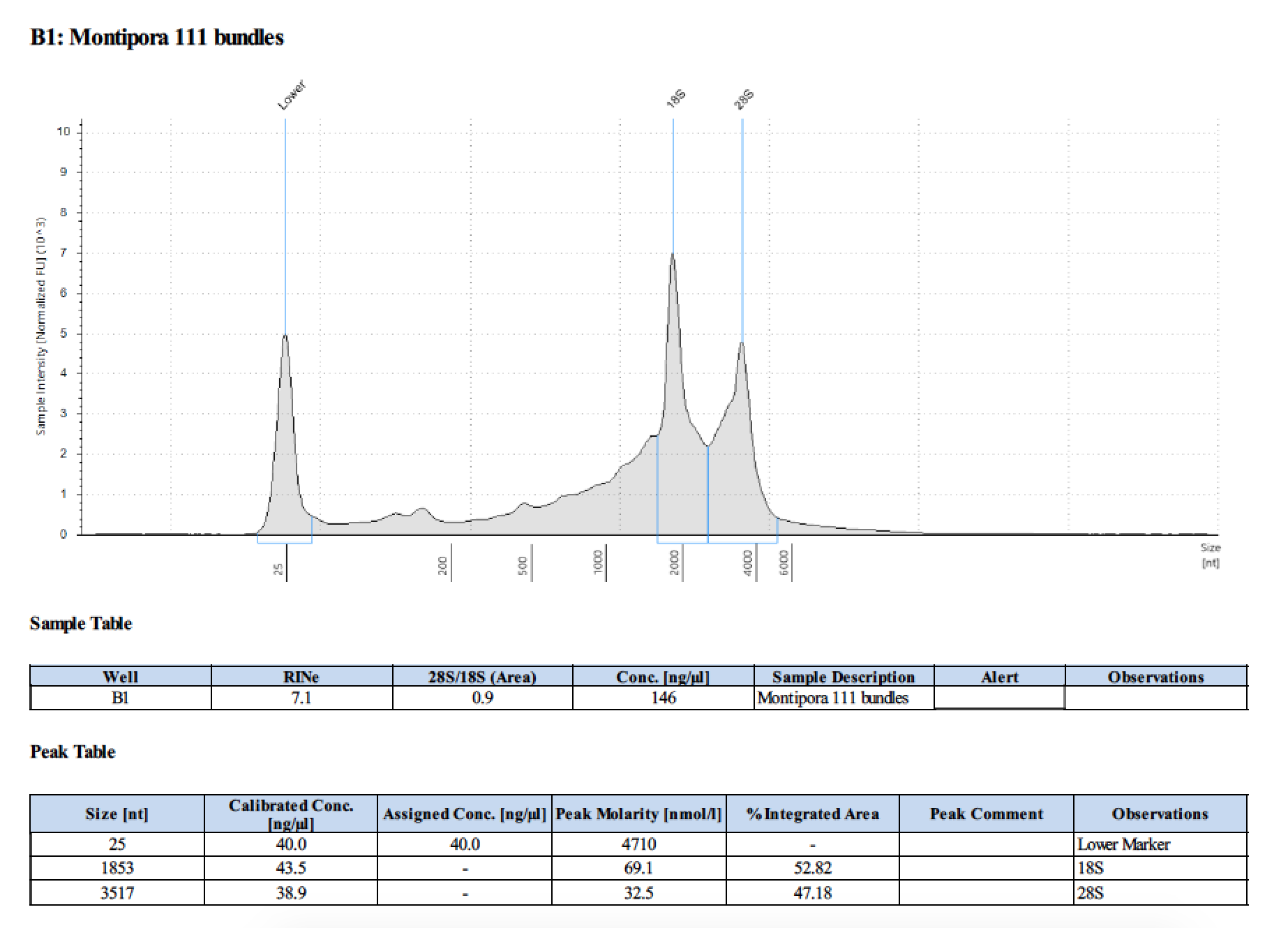
| 111 | 2018/06/13 | bundles | double DNA/RNA recommended Shield vol |
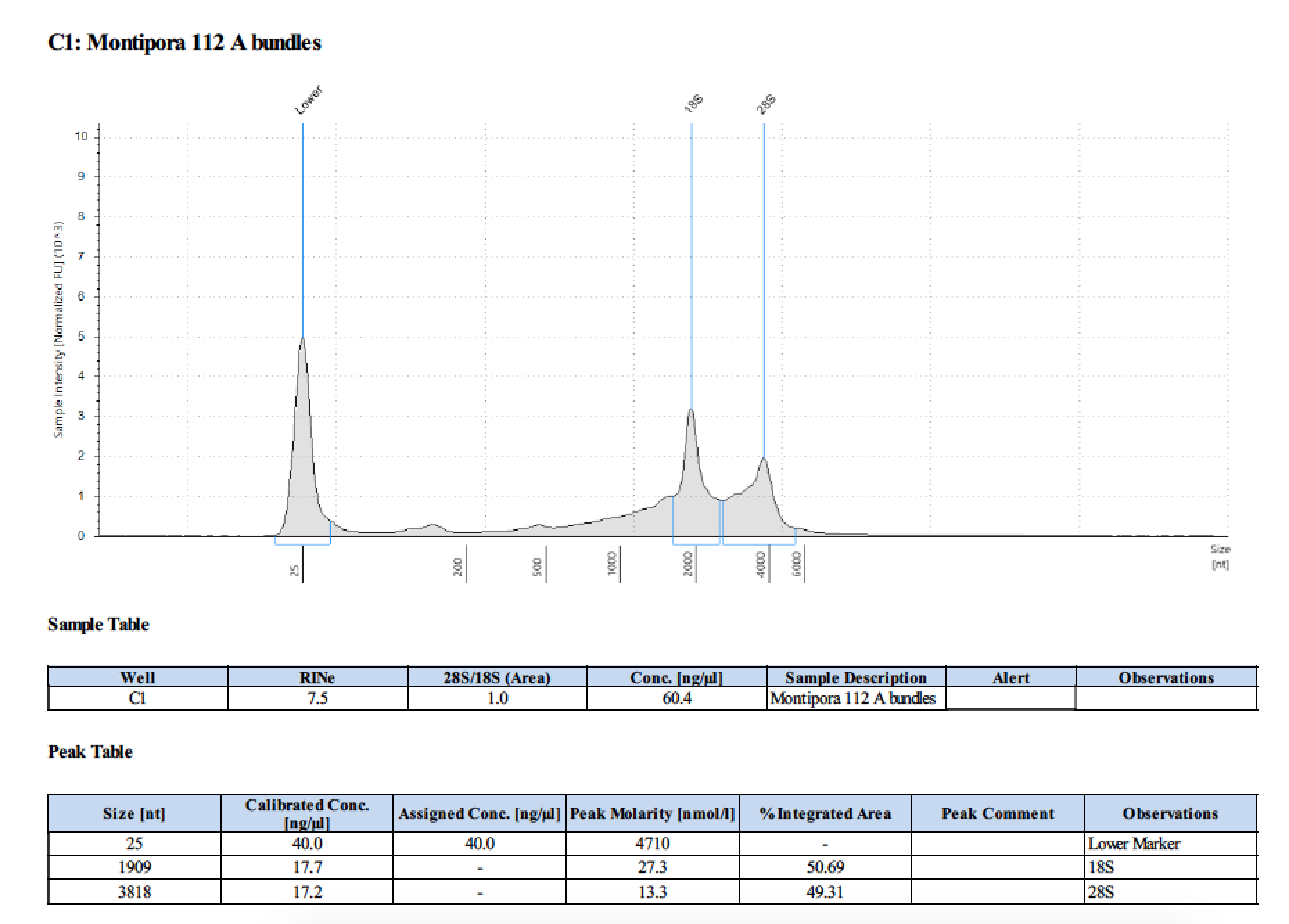
| 112 A | 2018/06/13 | bundles | double DNA/RNA Shield recommended vol and split into 2 tubes |
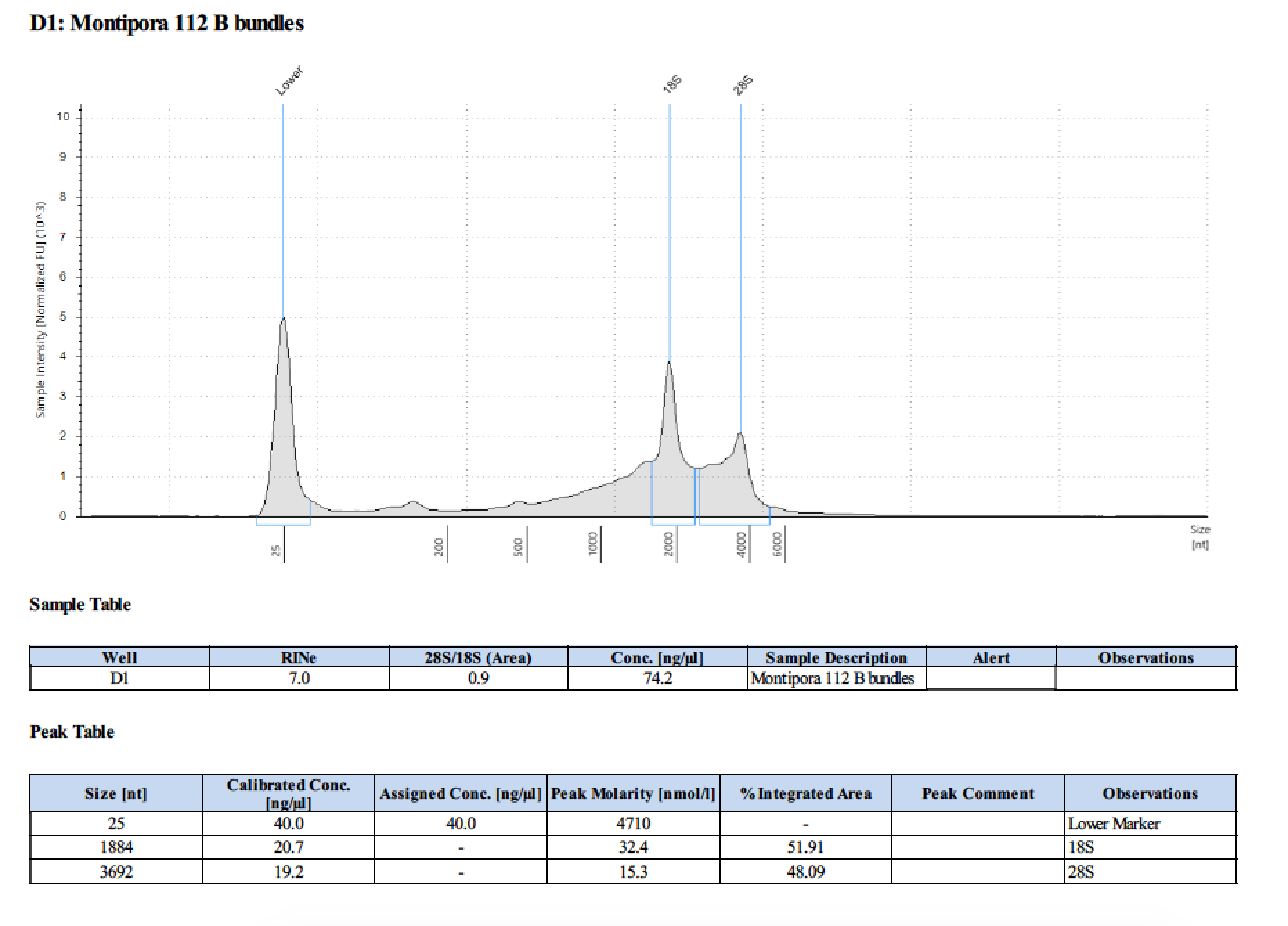
| 112 B | 2018/06/13 | bundles | double DNA/RNA Shield recommended vol and split into 2 tubes |
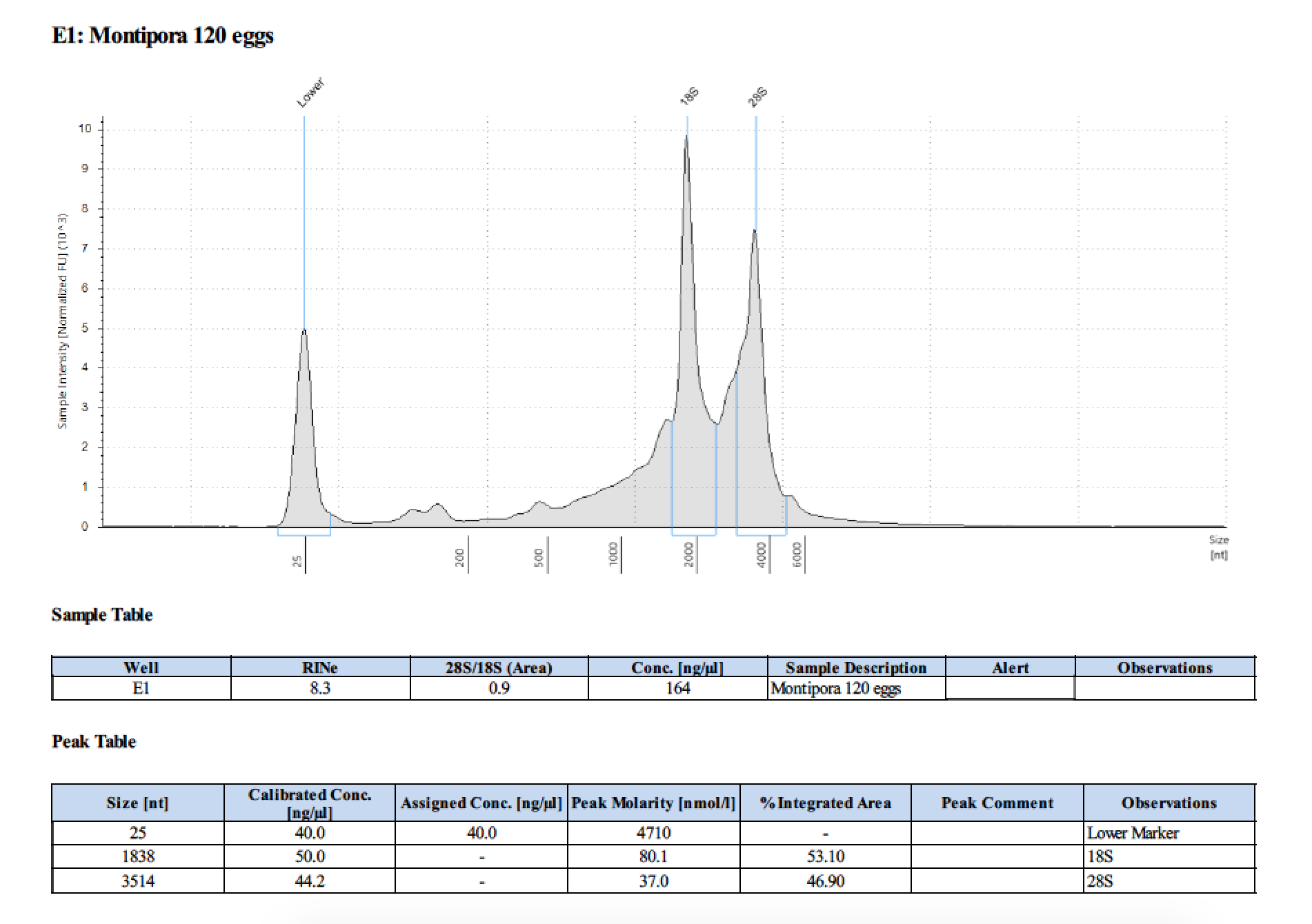
| 120 | 2018/06/13 | eggs | double DNA/RNA Shield recommended vol |
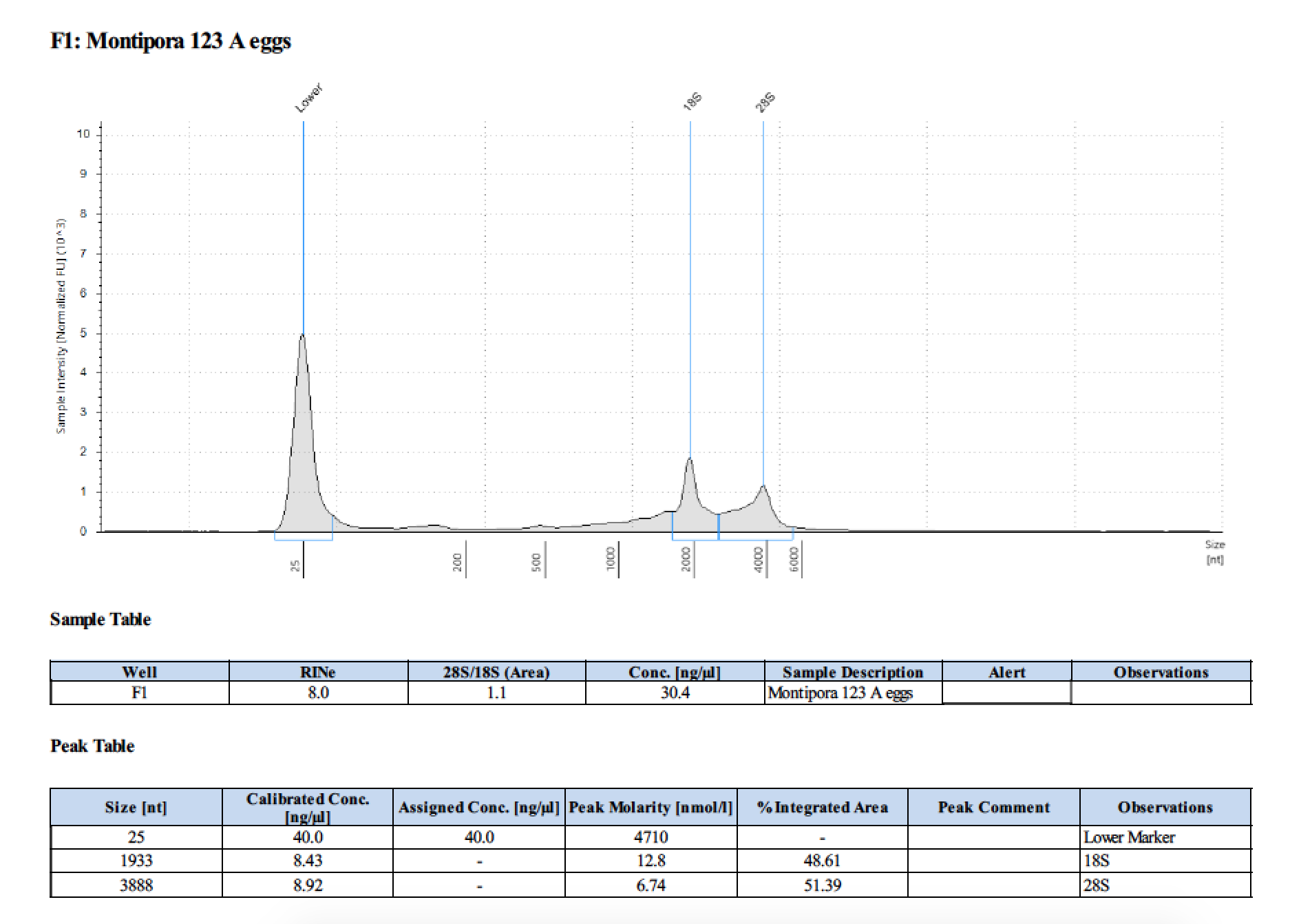
| 123 A | 2018/06/13 | eggs | double DNA/RNA Shield recommended vol and split into 2 tubes |
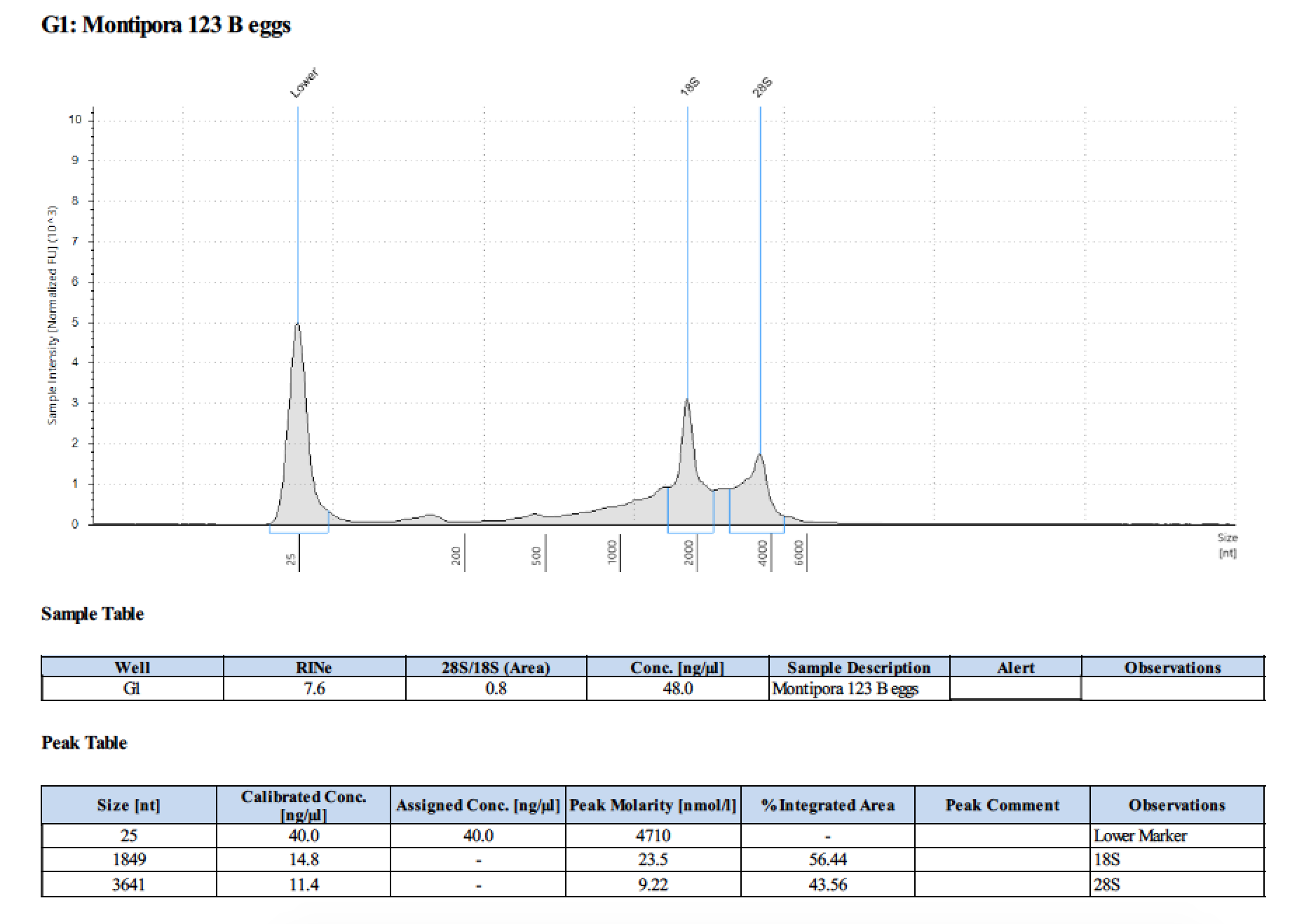
| 123 B | 2018/06/13 | eggs | double DNA/RNA Shield recommended vol and split into 2 tubes |
Mo’orea Samples
Trying two species: Massive Porites and Pocillopora verrucosa. Samples are stored in the -20 in DNA/RNA Shield. For some reason 1 sample of each species is liquid at -20 and one sample of each species is frozen. To test, I tried taking just the liquid from the liquid samples and bead homogenizing with the frozen samples.
| Sample # | Date Collected | Type | Extraction method |
|---|---|---|---|
| 1 | 2018/03/11 | Massive Porites | take liquid ~250µl |
| 2 | 2018/03/11 | Massive Porites | bead homogenize |
| 11 | 2018/03/11 | Pocillopora verrucosa | take liquid ~250µl |
| 12 | 2018/03/11 | Pocillopora verrucosa | bead homogenize |
Sample Prep
Eggs and Bundles:
- Samples were taken 1 at a time out of the -80 to minimize thawing
- 600µl of DNA/RNA Shield was added to each tube
- Half of the volume was transferred to second tubes for samples 112 and 123
It was very hard to pipette up the eggs/bundles even with the p1000. They had probably begun to lyse, sometimes the pipette got clogged or it was mucus-y - For samples 111 and 120 60µl of PK Digestion buffer and 30µl of Proteinase K were added
- For samples 112 A, 112 B, 123 A, and 123 B, 30µl of PK Digestion buffer and 15µl of Proteinase K were added
- All samples were vortexed and spun down on the table-top mini centrifuge
All samples were tough to get all the eggs/bundles to the bottom of the tube submerged in the liquid. They float and stick to the walls of the tube - Samples were put in the thermomixer at 55 degrees C shaking at 1200 at 10:15am
Mo’orea Samples:
- For samples 1 and 11, they were already liquid when taken out of the -20
- Most of the liquid was aspirated off (~250µl) and placed into a new tube. The remaining tissue was returned to the -20
- For samples 2 and 12, they were thawed and beads were poured into the tubes. They were homogenized by vortexing for ~30 seconds
- About ~250µl of liquid was able to be taken from the tubes and was transferred to new tubes
DNA Extraction
- Eggs and bundles samples were taken out of the thermomixer at 12:45pm. There was some undigested tissue that would not digest, mostly in samples 111 and 120. All tubes were spun down and the supernatant transferred to new tubes. 111 and 120 were transferred to 5mL tubes
- Equal volumes of DNA/RNA Lysis Buffer were added to every sample tube:
| Sample # | Volume added |
|---|---|
| 111 | 600µl |
| 112 A | 300µl |
| 112 B | 300µl |
| 120 | 600µl |
| 123 A | 300µl |
| 123 B | 300µl |
| 1 | 250µl |
| 2 | 250µl |
| 11 | 250µl |
| 12 | 250µl |
- Mix samples by flicking and spinning down
- 600µl of sample was gently added to Yellow DNA spin columns
- Centrifuged columns at 16000 rcf for 30 seconds
- Flow-through was transferred to new 1.5 or 5mL tubes labeled for RNA
- Samples 111 and 120 had an additional 600µl of sample that was spun through the column and the flow-through saved
note: samples 111 and 120 had white flakes/blobs that went through the DNA column and stuck to the sides of the collection tube, even though I tried to not aspirate any un-digested tissue after lysis. Samples 1 and 2 needed new collection tubes after their initial spin because a pellet (symbionts?) formed at the bottom of the collection tube
Additional steps follow this extraction procedure
- Add 400µl DNA/RNA Prep Buffer gently to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 700µl DNA/RNA Wash Buffer gently to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 400µl DNA/RNA Wash Buffer genetly to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 2 minutes
- Discard flow through (Zymo kit waste)
- Transfer yellow columns to new 1.5mL microcentrifuge tubes
- Add 50µl warmed 10mM Tris HCl to each yellow DNA column by dripping slowly directly on the filer
- Incubate at room temp for 5 minutes
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Repeat last three steps for a final elution volume of 100µl
- Label tubes, store at 4 degrees C if quantifying the same day or the next, if waiting longer store in -20
RNA Extraction
- Add equal volume 100% EtOH to the 1.5mL and 5mL tubes labeled for RNA containing the original yellow column flow through
| Sample # | Volume added |
|---|---|
| 111 | 1200µl |
| 112 A | 600µl |
| 112 B | 600µl |
| 120 | 1200µl |
| 123 A | 600µl |
| 123 B | 600µl |
| 1 | 500µl |
| 2 | 500µl |
| 11 | 500µl |
| 12 | 500µl |
- Vortex and spin down to mix
- Add 700µl of that liquid to the green RNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 700µl to the green RNA spin columns (the rest from the 1.5mL RNA tubes)
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 400µl DNA/RNA Wash Buffer gently to each green RNA column
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Make DNase I treatment master mix:
- 75µl DNA Digestion buffer x # of samples
- 5µl DNase I x # of samples
- Add 80µl DNase I treatment master mix directly to the filter of the green RNA columns
- Incubate at room temp for 15 minutes
- Add 400µl DNA/RNA Prep Buffer gently to each column
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 700µl DNA/RNA Wash Buffer gently to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Discard flow through (Zymo kit waste)
- Add 400µl DNA/RNA Wash Buffer genetly to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 2 minutes
- Discard flow through (Zymo kit waste)
- Transfer green columns to new 1.5mL microcentrifuge tubes
- Add 50µl warmed DNase/RNase free water to each green RNA column by dripping slowly directly on the filer
- Incubate at room temp for 5 minutes
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Repeat last three steps for a final elution volume of 100µl
- Label 1.5mL tubes on ice afterwards, and aliquot 5µl into PCR strip tubes to save for Qubit and Tape Station to avoid freeze-thaw of your stock sample
- Store all tubes in the -80
Qubit
- Broad Range dsDNA and RNA Qubit protocol
- All samples were read twice
| Sample | DNA Standard 1 (RFU) | DNA Standard 2 (RFU) | DNA 1 (ng/µl) | DNA 2 (ng/µl) | Average DNA | RNA Standard 1 (RFU) | RNA Standard 2 (RFU) | RNA 1 (ng/µl) | RNA 2 (ng/ul) | Average RNA |
|---|---|---|---|---|---|---|---|---|---|---|
| 111 | 199.2 | 22813 | 72.8 | 74.2 | 73.5 | 412 | 11540 | 171 | 170 | 170.5 |
| 112 A | 199.2 | 22813 | 53.8 | 54.0 | 53.9 | 412 | 11540 | 56.8 | 56.6 | 56.7 |
| 112 B | 199.2 | 22813 | 42.2 | 42.4 | 42.3 | 412 | 11540 | 69.8 | 69.2 | 69.5 |
| 120 | 199.2 | 22813 | too low | - | - | 412 | 11540 | 169 | 168 | 168.5 |
| 123 A | 199.2 | 22813 | too low | - | - | 412 | 11540 | 28.4 | 28.2 | 28.3 |
| 123 B | 199.2 | 22813 | too low | - | - | 412 | 11540 | 51.0 | 50.6 | 50.8 |
| 1 | 199.2 | 22813 | 7.94 | 8.02 | 7.98 | 412 | 11540 | too low | - | - |
| 2 | 199.2 | 22813 | 9.92 | 9.92 | 9.92 | 412 | 11540 | 22.6 | 22.4 | 22.5 |
| 11 | 199.2 | 22813 | 8.38 | 8.34 | 8.36 | 412 | 11540 | 12.0 | 11.6 | 11.8 |
| 12 | 199.2 | 22813 | 24.2 | 24.4 | 24.4 | 412 | 11540 | 29.0 | 28.8 | 28.9 |
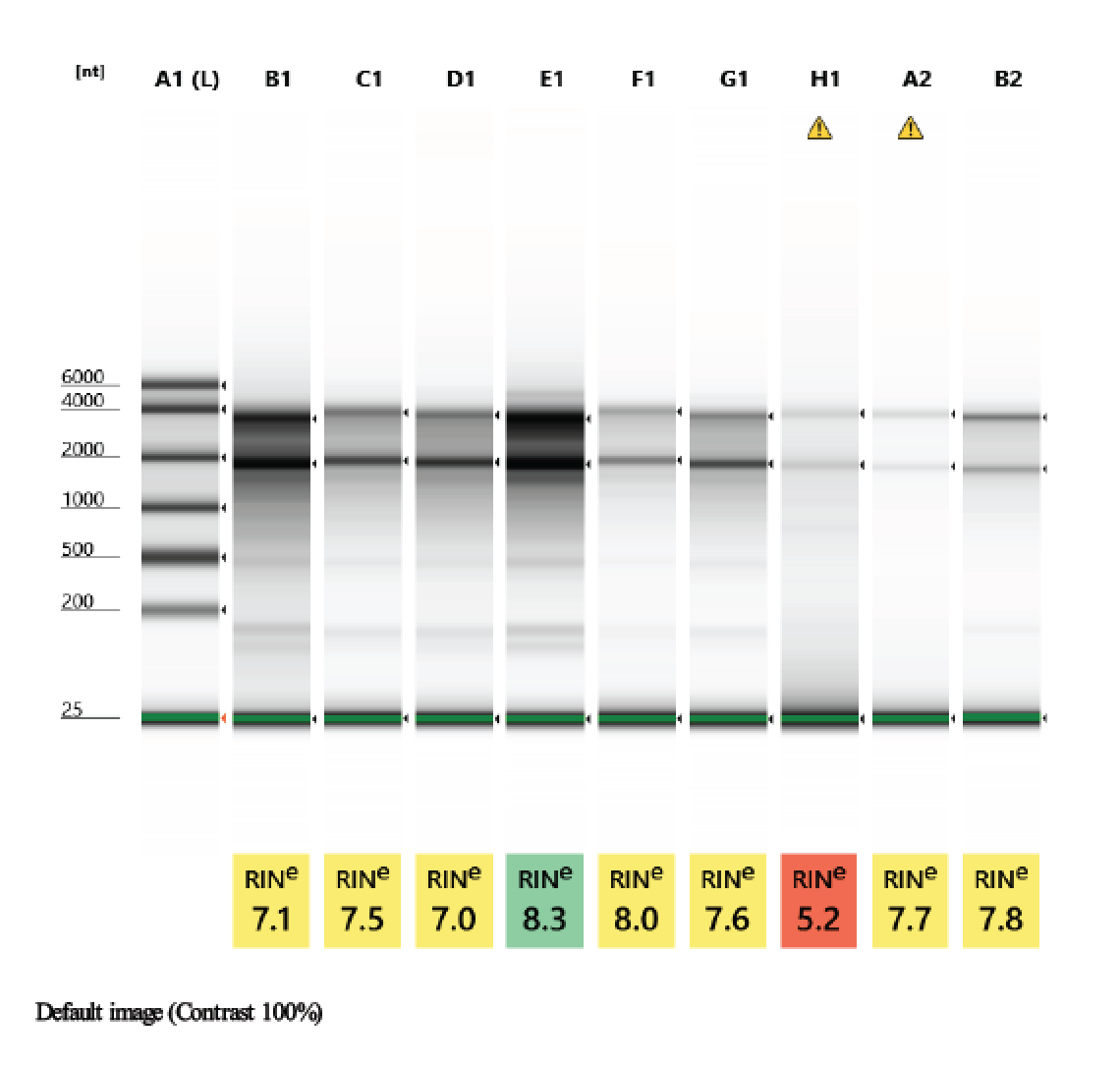
TapeStation
- Followed RNA TapeStation protocol
- Did not run sample 1 because there was no RNA
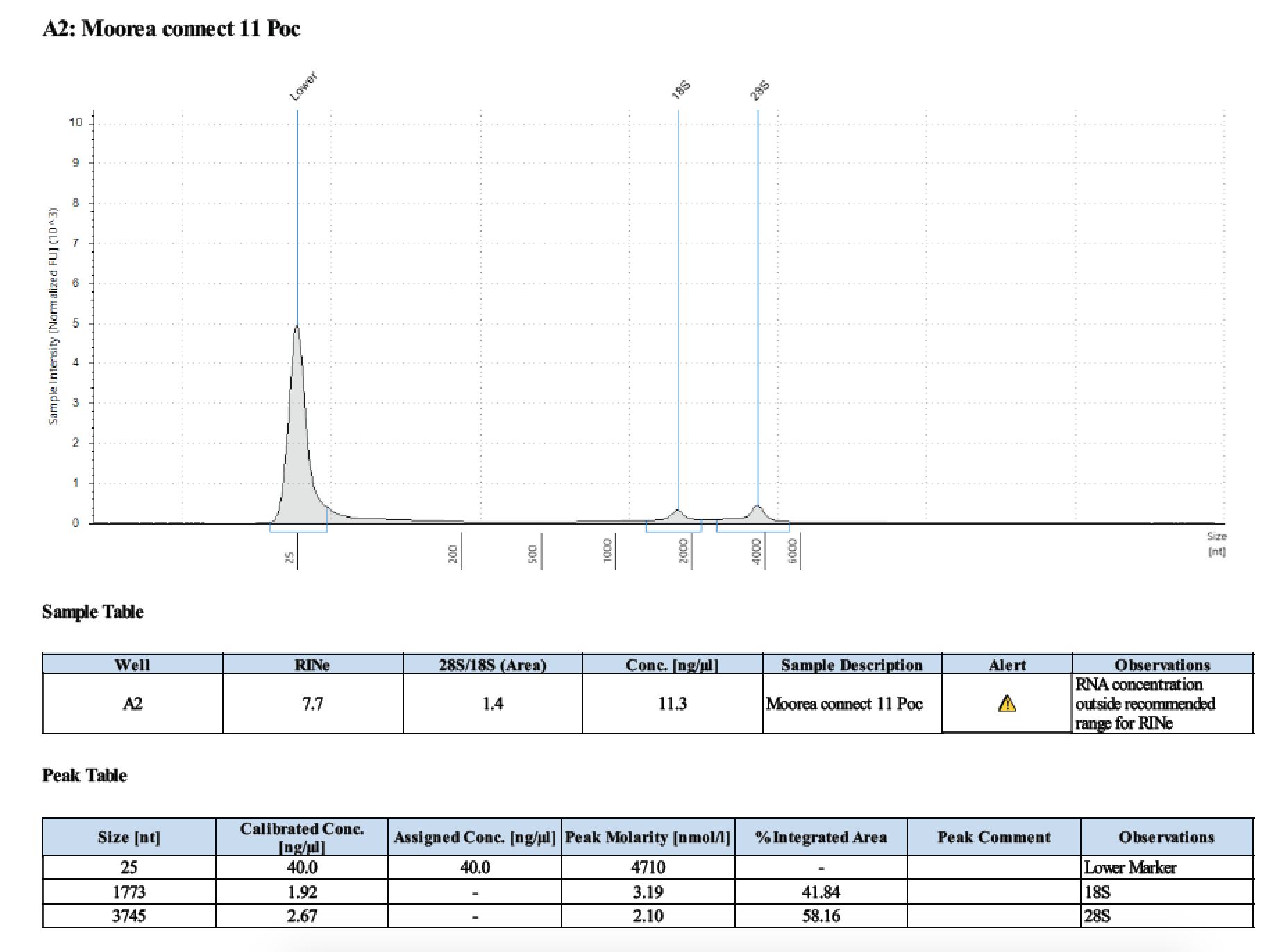
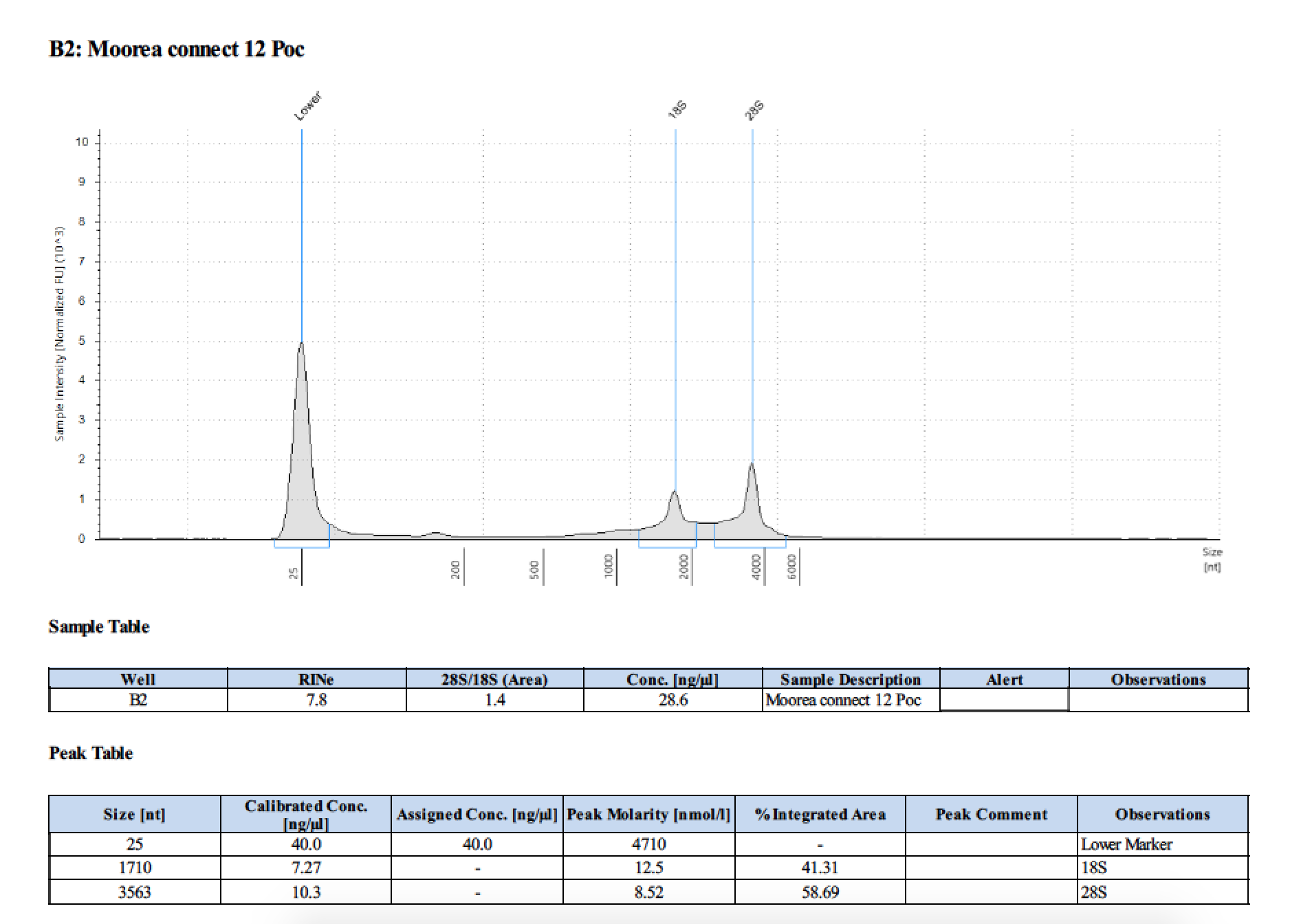
Written on March 17, 2019