Dilution DNA to 2ng/ul from Electroporation Tests and Redoing PCRs
It could be that the DNA I am using for the electroporation tests to see if there is on DiNV DNA left in the electroporated samples is too concentrated and giving me false or missleading results.
Using samples from this and this tests.
First I Qubited each sample and then diluted it to 2ng/ul using the qubit protocol
| sample | ng/ul | vol DNA for 2ng/ul | vol hydration solution for 2ng/ul |
|---|---|---|---|
| 8A | 12.6 | 3.33ul | 16.67ul |
| 9A | 8.81 | 4.54ul | 15.46ul |
| 10A | 9.27 | 4.3ul | 15.7ul |
| 11A | 6.89 | 2.9ul | 7.1ul |
| 8B | 7.51 | 2.66ul | 7.34ul |
| 9B | 13.1 | 3.05ul | 17ul |
| 10B | 6.96 | 2.87ul | 7.13ul |
| 11B | 8.86 | 4.6ul | 15.4ul |
| 12A | 7.98 | 2.5ul | 7.5ul |
| 13A | 6.95 | 2.88ul | 7.12 |
| 12B | 2.48 | 5.64ul | 1.36ul |
| 13B | 4.11 | 4.87ul | 5ul |
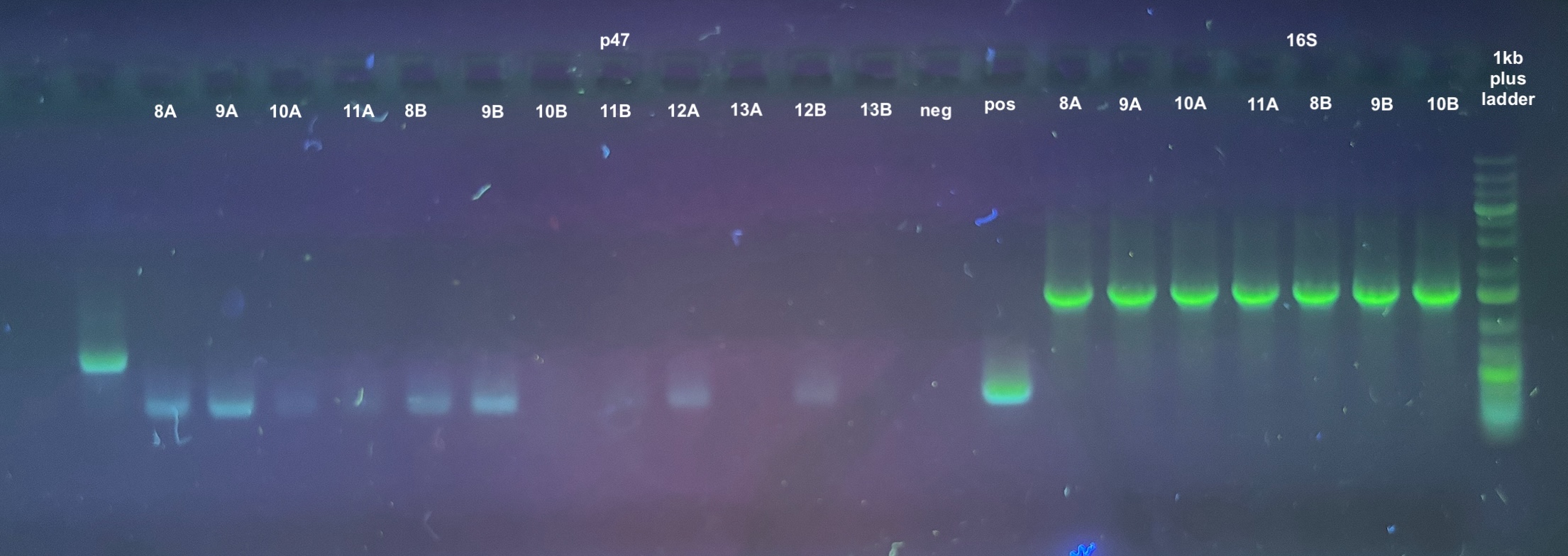
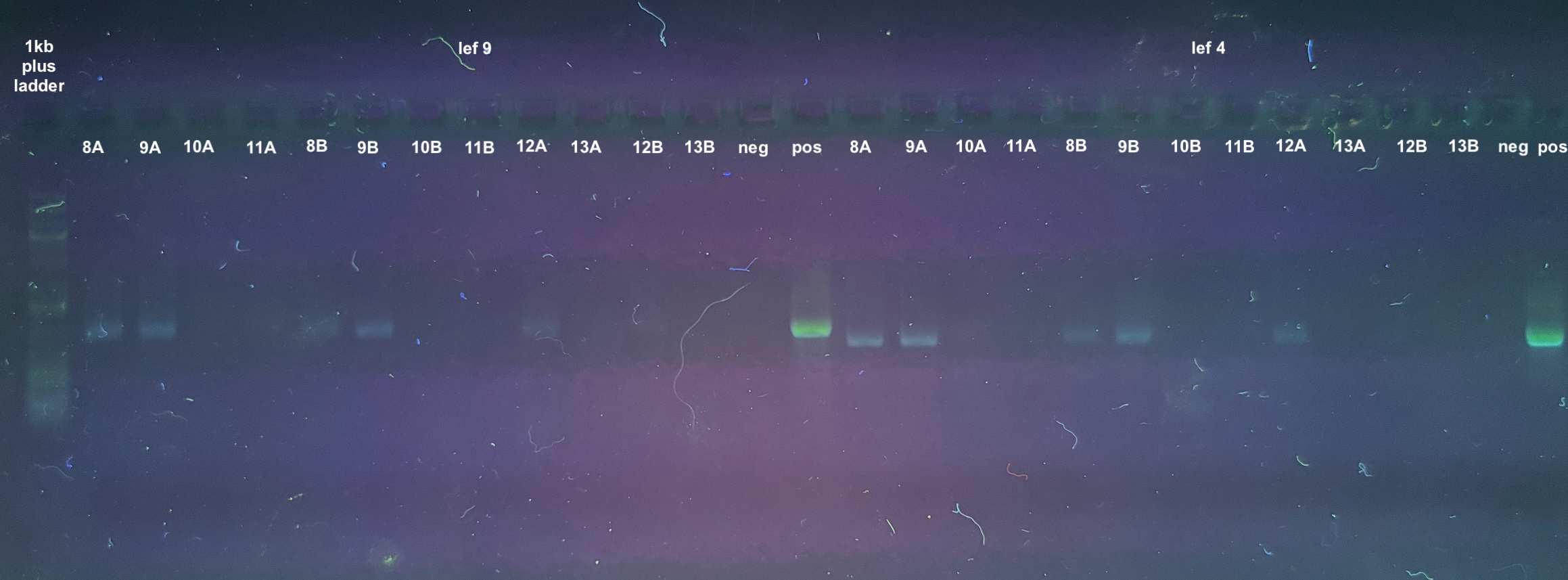
PCRs
- 4 PCRs were run on the samples, and the process followed the general PCR protocol completely. Master mix volumes are listed here:
| reagent | p47 | 16S | lef 9 | lef 4 |
|---|---|---|---|---|
| GoTaq | 75ul | 75ul | 75ul | 75ul |
| F primer | 3.75ul | 3.75ul | 3.75ul | 3.75ul |
| R primer | 3.75ul | 3.75ul | 3.75ul | 3.75ul |
| molecular grade water | 52.5ul | 52.5ul | 52.5ul | 52.5ul |
- All PCR programs were run for 30 cycles, and program information can be found here
- 1% gels were run at 90V for 45 minutes to resolve the bands: