Suite of PCR Reactions and Preparation for Sanger Sequencing of Fly Homogenate and Week 9 Experimental Evolution DNA Extracts
Using all the experimental evolutions primers that are designed around interesting SNPs on the fly homogenate and week 9 sampling, extracted here.
20231206 PCRs
All DNA samples were diluted 1:1 in DNA hydration solution before use in these PCRs. I used samples: 43, 50, 58, and 41. See samples information here. All samples, primers, reagents, mixes, and reactions were kept on ice. Everything was thawed on ice, vortexed, and spun down before use.
There were 4 samples plus a positive and negative control for each reaction. All reaction mixes had the same amount of components, except different primers. Information on the primers and their PCR programs can be found here. Note that all PCR reactions were run for 33 cycles, and that the 40-1 PCR was done twice and increased to 35 cycles because of poor amplification.
Reaction mixes:
- 32.5ul GoTaq
- 1.625ul F primer
- 1.625ul R primer
- 22.7ul molecular grade water
The gel was a 1% gel run for 55 minutes at 80V (this was too long and one of the products left the gel rig). I only used 3ul of PCR product for the gel because I wanted to save as much as I could for the sequencing.
First gel is all primers (Last one left the gel)
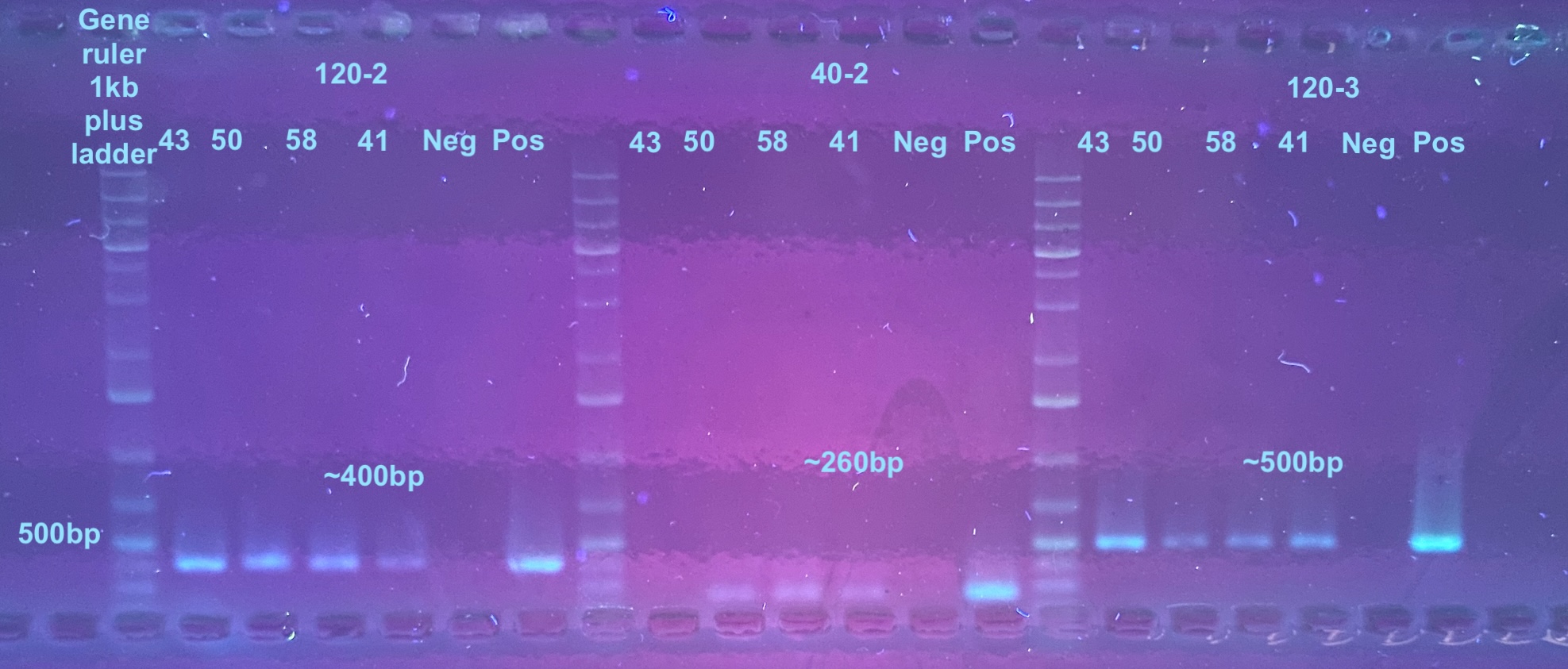
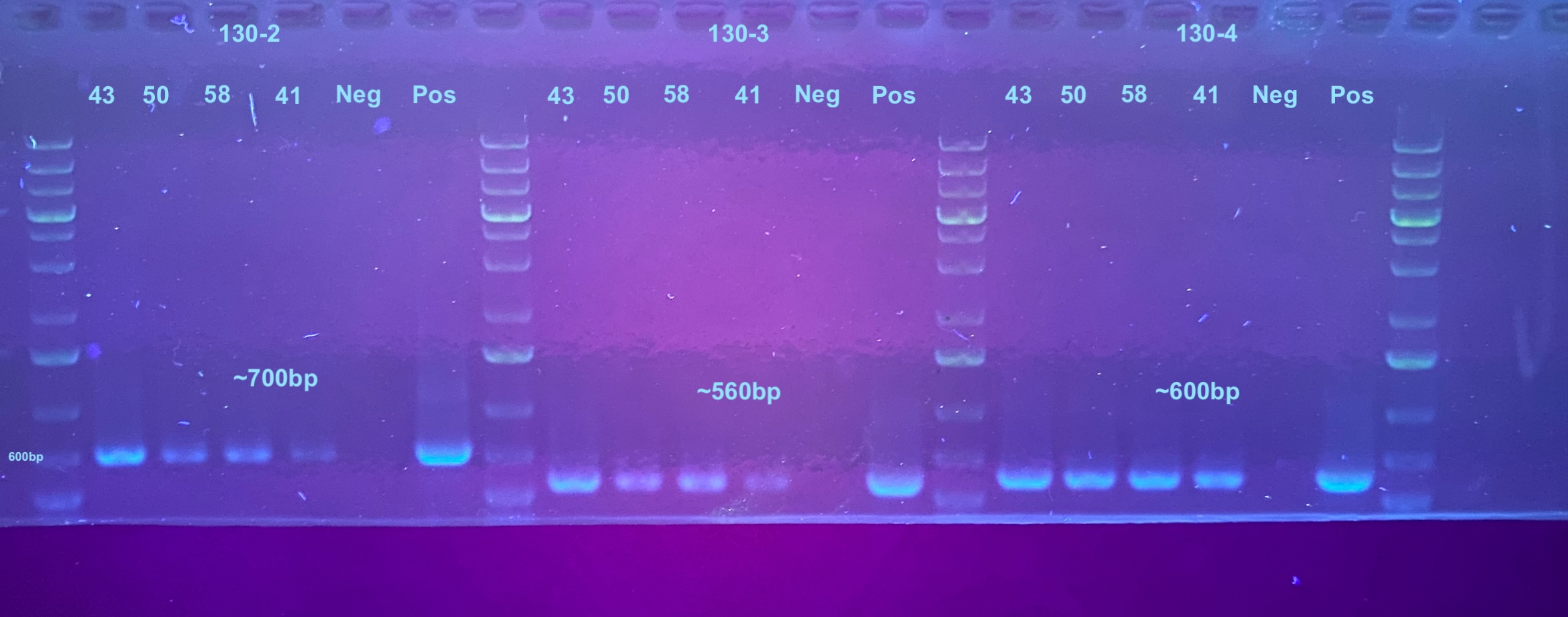
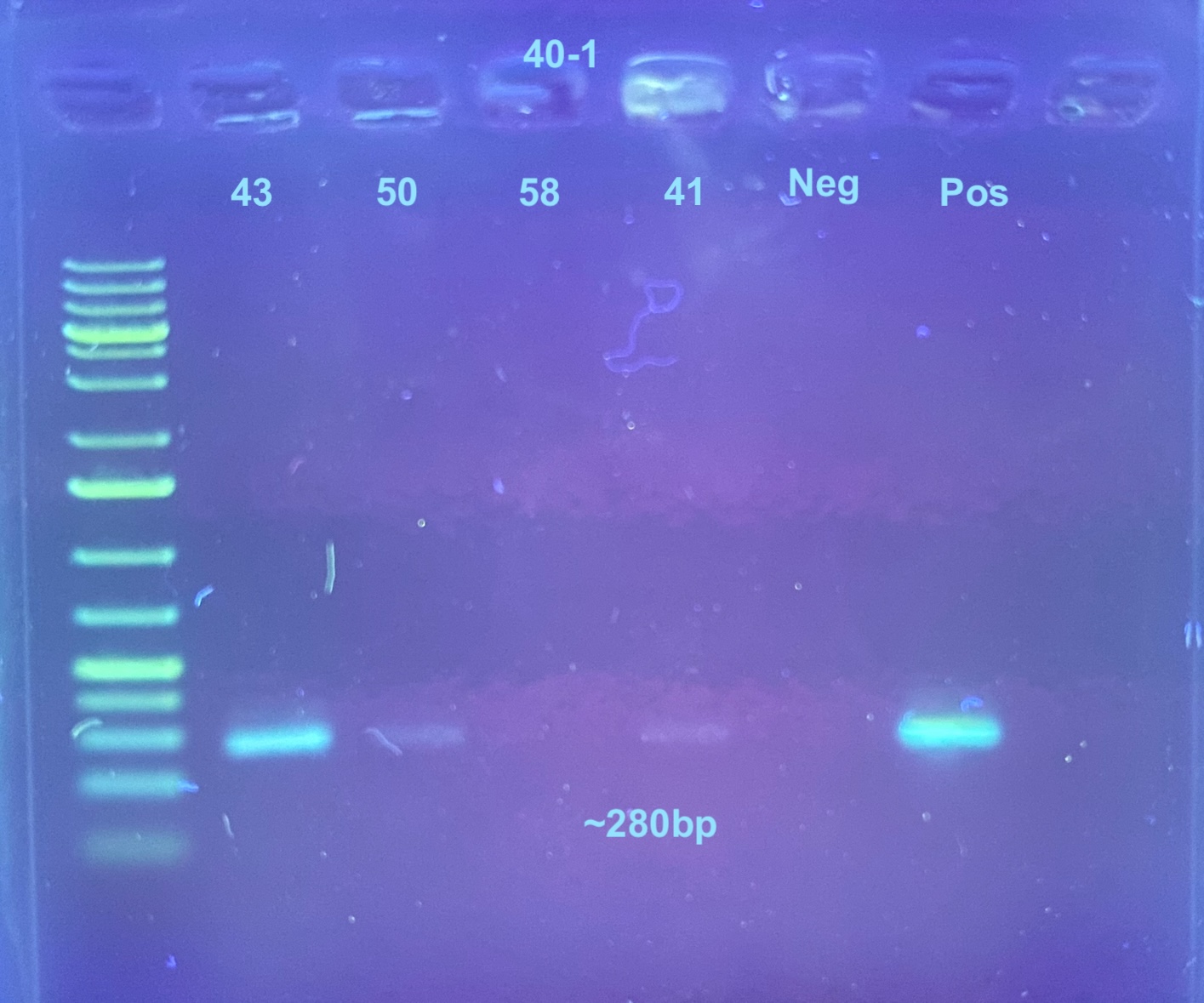
Second gel is just 40-1 re-do PCR with 35 cycles done 2 days later
20231207 Bead Clean and Qubit
All samples were prepared the same way (even the 40-1 samples that were re-done)
- Took Ampure beads out of fridge and swirled to resuspend and get to room temperature
- Made fresh 80% ethanol
- Added 8ul of molecular grade water to each PCR reaction (samples 43, 50, 58, and 41 from each primer)
- There should be 7ul left from the gel, giving me a total of 15ul after adding the water
- Added 1.5X beads - 22.5ul of beads to each sample and pipette mixed
- Let samples incubate on the orbital shaker for 15 minutes
- Transferred samples to the magnet plate and let the beads collect
- Removed ~34ul of supernatant and discarded
- Added 100ul of fresh 80% ethanol to each tube without disturbing the beads
- Removed the ethanol without disturbing the beads
- Added another 100ul of 80% ethanol to each tube without disturbing the beads
- Removed the ethanol without disturbing the beads
- Removed last remaining ethanol with 10ul
- Added 16ul of molecular grade water off the magnet to each tube and pipetted to resuspend the bead pellet
- Let tubes incubate on the orbital shaker for 5 minutes
- Placed tubes back on the magnet
- Removed 15ul of the clear supernatant to new tubes labeled 1-28
Each sample was run on the high sensitivity Qubit to quantify DNA following this protocol. Notice that the 40-1 samples were done twice.
| tube number | sample | primer | qubit |
|---|---|---|---|
| 1 | 43 | 120-2 | 10.8 |
| 2 | 50 | 120-2 | 4.57 |
| 3 | 58 | 120-2 | 5.11 |
| 4 | 41 | 120-2 | 2.87 |
| 5 | 43 | 40-2 | 0.17 |
| 6 | 50 | 40-2 | 3.02 |
| 7 | 58 | 40-2 | 3.27 |
| 8 | 41 | 40-2 | 1.97 |
| 9 | 43 | 120-3 | 7.85 |
| 10 | 50 | 120-3 | 2.08 |
| 11 | 58 | 120-3 | 2.45 |
| 12 | 41 | 120-3 | 1.6 |
| 13 | 43 | 130-2 | 4.37 |
| 14 | 50 | 130-2 | 1.22 |
| 15 | 58 | 130-2 | 1.42 |
| 16 | 41 | 130-2 | 0.614 |
| 17 | 43 | 130-3 | 6.4 |
| 18 | 50 | 130-3 | 1.82 |
| 19 | 58 | 130-3 | 2.32 |
| 20 | 41 | 130-3 | 0.998 |
| 21 | 43 | 130-4 | 9.64 |
| 22 | 50 | 130-4 | 4.3 |
| 23 | 58 | 130-4 | 4.64 |
| 24 | 41 | 130-4 | 3.22 |
| 25 | 43 | 40-1 | 1.21 |
| 26 | 50 | 40-1 | 0.13 |
| 27 | 58 | 40-1 | 0.14 |
| 28 | 41 | 40-1 | too low |
| 25-2 | 43 | 40-1 | 4.36 |
| 26-2 | 50 | 40-1 | 0.758 |
| 27-2 | 58 | 40-1 | too low |
| 28-2 | 41 | 40-1 | 0.246 |
Samples were put into the freezer until ready to be prepared.
20231210 Prep Samples for Sequencing
- Samples were thawed on ice
- Forward primers were thawed on ice
- Everything was vortexed and spun down before use
- I determined that for each sample ACGT recommends either 6-10ng or 10-20ng of DNA based on the size (0.6 to 2ng/ul). So I decided to do 10ng for every sample if I could, and for the 40-1 samples I brought it down to 6ng. Not every sample had enough so for some I just added the maximum amount 10ul and would just go with it
- ACGT wants 10ul of DNA and 2ul of primer
- For each of these I added the molecular grade water first, then the DNA, then the primer
- Every sample was put into 1.5mL tubes
- Sample tubes were parafilmed to protect in shipping
- Samples were sent out the next day for sequencing
| tube number | product size | DNA needed in 10ul | ul for 10ng | ul to 10ul | ul primer | primer |
|---|---|---|---|---|---|---|
| 1 | 400bp | 6-10ng | 0.93 | 9.07 | 2 | 120-2 |
| 2 | 400bp | 6-10ng | 2.19 | 7.81 | 2 | 120-2 |
| 3 | 400bp | 6-10ng | 1.96 | 8.04 | 2 | 120-2 |
| 4 | 400bp | 6-10ng | 3.48 | 6.52 | 2 | 120-2 |
| 5 | 261bp | 6-10ng | 10.00 | 0.00 | 2 | 40-2 |
| 6 | 261bp | 6-10ng | 3.31 | 6.69 | 2 | 40-2 |
| 7 | 261bp | 6-10ng | 3.06 | 6.94 | 2 | 40-2 |
| 8 | 261bp | 6-10ng | 5.08 | 4.92 | 2 | 40-2 |
| 9 | 503bp | 10-20ng | 1.27 | 8.73 | 2 | 120-3 |
| 10 | 503bp | 10-20ng | 4.81 | 5.19 | 2 | 120-3 |
| 11 | 503bp | 10-20ng | 4.08 | 5.92 | 2 | 120-3 |
| 12 | 503bp | 10-20ng | 6.25 | 3.75 | 2 | 120-3 |
| 13 | 700bp | 10-20ng | 2.29 | 7.71 | 2 | 130-2 |
| 14 | 700bp | 10-20ng | 8.20 | 1.80 | 2 | 130-2 |
| 15 | 700bp | 10-20ng | 7.04 | 2.96 | 2 | 130-2 |
| 16 | 700bp | 10-20ng | 10.00 | 0.00 | 2 | 130-2 |
| 17 | 558bp | 10-20ng | 1.56 | 8.44 | 2 | 130-3 |
| 18 | 558bp | 10-20ng | 5.49 | 4.51 | 2 | 130-3 |
| 19 | 558bp | 10-20ng | 4.31 | 5.69 | 2 | 130-3 |
| 20 | 558bp | 10-20ng | 10.00 | 0.00 | 2 | 130-3 |
| 21 | 599bp | 10-20ng | 1.04 | 8.96 | 2 | 130-4 |
| 22 | 599bp | 10-20ng | 2.33 | 7.67 | 2 | 130-4 |
| 23 | 599bp | 10-20ng | 2.16 | 7.84 | 2 | 130-4 |
| 24 | 599bp | 10-20ng | 3.11 | 6.89 | 2 | 130-4 |
| 25 | 279bp | 6-10ng | NA | NA | 2 | 40-1 |
| 26 | 279bp | 6-10ng | NA | NA | 2 | 40-1 |
| 27 | 279bp | 6-10ng | 10.00 | 0.00 | 2 | 40-1 |
| 28 | 279bp | 6-10ng | NA | NA | 2 | 40-1 |
| 25-2 | 279bp | 6-10ng | 1.38 | 8.62 | 2 | 40-1 |
| 26-2 | 279bp | 6-10ng | 7.92 | 2.08 | 2 | 40-1 |
| 27-2 | 279bp | 6-10ng | NA | NA | 2 | 40-1 |
| 28-2 | 279bp | 6-10ng | 10.00 | 0.00 | 2 | 40-1 |