Designing Primers around SNPs Potentially Important for DiNV Adaptation to Cell Culture and Testing the Primers
Designing Primers
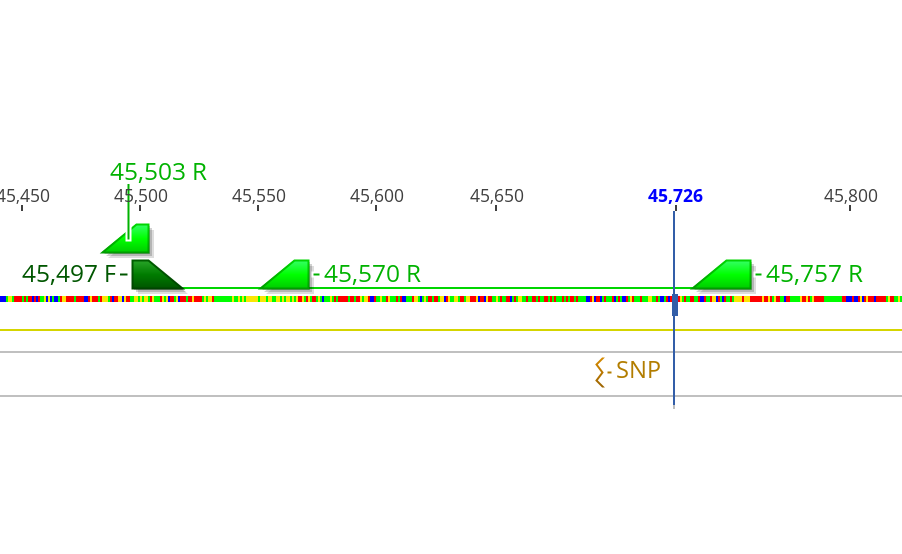
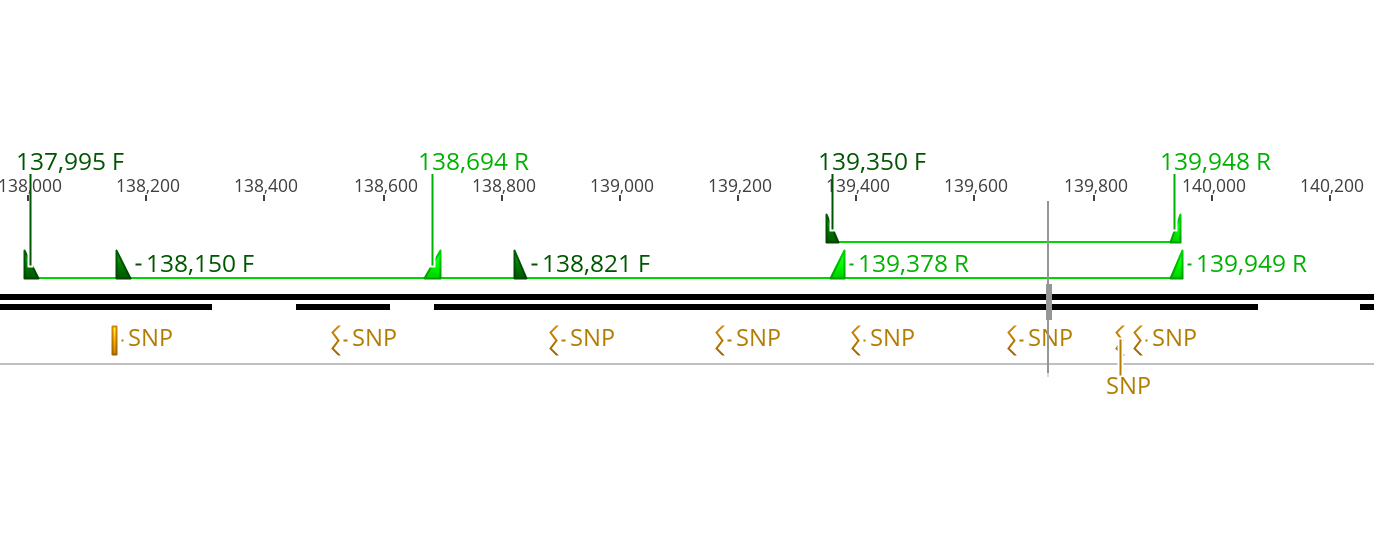
- The SNPs the primers were designed around to catch are these which were identified to be significantly associated differences in both the Gprime and the Fisher’s Exact test methods for looing at evolution
- See here for fisher’s exact test and Gprime analysis
- Additionally 3 SNPs: 42,865, 43,049, and 45,695 which were fixed different between the two samples and were in a minor but not significant peak in the Gprime analysis were used to base primers around
- I used Geneious software to design the primers, I generated large primers (1000bp plus products) for the 120kb and 130kb regions, as well as smaller ~200-500bp primer sets. The idea would be that not all SNPs need to be covered, but that a good amount should. Not all primers might work, it was hard to design primers that fit all the criteria of making a good primer, and larger products are harder to amplify
- All primer specifics can be found in this document
- And the locations of the primers in the genome relative to the SNPs can be see in the pictures below:


Testing Primers 20231102 and 20231103
All primers were diluted to 100uM concentration with low EDTA TE buffer for the stock concentration, and further diluted to 10uM for the working solution concentration.
Pretty much all the primer pairs have different TMs or expected product sizes, so only 2 out of the 9 sets I started out with on the sample PCR program. All PCR programs tested can be found here. All programs were made to cycle 30 times.
For each pair of primers to test, I used the same 3 samples, samples 2 and 5 from the clarification pellet hirt extraction diluted 1:50, and sample 29 dead infected fly DNA diluted to 1ng/ul. I also used a negative control with water.
Each master mix included:
- 22.5ul GoTaq
- 1.125ul Forward primer 10uM
- 1.125ul Reverse primer 10uM
- 15.75ul molecular grade water
The master mixes, DNA, and PCR tubes were all made and kept on ice. Everything was vortexed and spun down before use.
After all PCRs were done, a large 1.5% gel was run for 50 minutes at 90V. 165mL 1X TAE, 2.475g agarose and 2ul Midori stain.
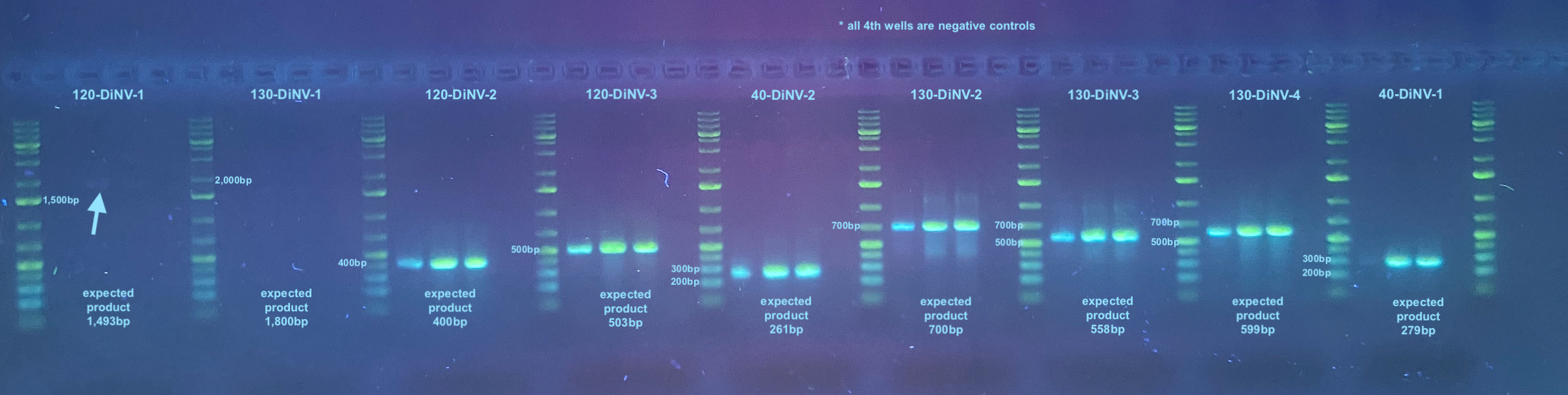
This shows that all primers except for the 1st two seem to work with no off target amplification. The first set, 120-DiNV-1 looks like it might have a little amplification at a size larger than it should be, but it is hard to tell. Those primers may be worked on in the future, or maybe not.