Taking Samples and Fluid Changing Flasks at the 9 Week Mark for Experimental DiNV Evolution in Dinn Cells (~63 days) and Extracting DNA from 1 50ul Sample Per Replicate Flask, and Checking Samples for Virus Presence
Sampling 20231107
Cells were inoculated on the 26th of Sept see post here. There are 3 replicate flasks that have been left to grow and get infected in the 23C incubator for 42 days. I am only removing supernatant for samples, the cells are not getting passed throughout this experiment, the only thing that happens is a tri-weekly fluid change (once every three weeks). In the six week sampling I took 7 50ul supernatant and 1 1mL supernatant sample per replicate flask, and I did the same style of sampling here as well.
All steps took place in the cell culture hood
- Made fresh 10% FBS, 4% mushroom Schneider’s medium with antibiotics in the hood to room temperature
- Prepared 1.5mL tubes to take samples
- Removed 7 samples of 50ul and 1 sample of 1000ul from each flask before changing fluid and placed into the 1.5mL tubes:
| tube number | day sampled | date sampled | sample volume | replicate tube | flask from |
|---|---|---|---|---|---|
| 42 | day 63 | 20231128 | 50ul | 1 | A |
| 43 | day 63 | 20231128 | 50ul | 2 | A |
| 44 | day 63 | 20231128 | 50ul | 3 | A |
| 45 | day 63 | 20231128 | 50ul | 4 | A |
| 46 | day 63 | 20231128 | 50ul | 5 | A |
| 47 | day 63 | 20231128 | 50ul | 6 | A |
| 48 | day 63 | 20231128 | 50ul | 7 | A |
| 49 | day 63 | 20231128 | 1mL | NA | A |
| 50 | day 63 | 20231128 | 50ul | 1 | B |
| 51 | day 63 | 20231128 | 50ul | 2 | B |
| 52 | day 63 | 20231128 | 50ul | 3 | B |
| 53 | day 63 | 20231128 | 50ul | 4 | B |
| 54 | day 63 | 20231128 | 50ul | 5 | B |
| 55 | day 63 | 20231128 | 50ul | 6 | B |
| 56 | day 63 | 20231128 | 50ul | 7 | B |
| 57 | day 63 | 20231128 | 1mL | NA | B |
| 58 | day 63 | 20231128 | 50ul | 1 | C |
| 59 | day 63 | 20231128 | 50ul | 2 | C |
| 60 | day 63 | 20231128 | 50ul | 3 | C |
| 61 | day 63 | 20231128 | 50ul | 4 | C |
| 62 | day 63 | 20231128 | 50ul | 5 | C |
| 63 | day 63 | 20231128 | 50ul | 6 | C |
| 64 | day 63 | 20231128 | 50ul | 7 | C |
| 65 | day 63 | 20231128 | 1mL | NA | C |
- Removed 1.5mL of remaining medium in each flask and discarded it into a 15mL conical. The conical was placed in the autoclave trash when full (should contain virus)
- Added 3mL of 10% FBS, 4% mushroom Schneider’s medium with antibiotics to each flask gently
- Gently rocked flasks to distribute the medium
- Placed flasks back in the 23C incubator for another 3 weeks
- Sample tubes were frozen at -20
Sample information can be found here
DNA Extractions 20231129
For these DNA extractions I added in a second tube of fly homogenate, and to balance everything (because it was an uneven number) I added a second replicate A extraction. For the fly homogenate, I thawed an aliquot (I think that had already been thawed and used once), removed 50ul to a sample tube, and froze what was left. I am not too concerned about the freeze thaws because the DNA should be relatively intact.
Samples:
| sample # | day | flask |
| 43 | day 63 | A |
| 50 | day 63 | B |
| 58 | day 63 | C |
All samples were thawed on ice, this general protocol was used, and filter tips were used at all times. Since all samples were presumed positive for DiNV, there was no specific order to them. 2 Extraction controls were done at the same time as well.
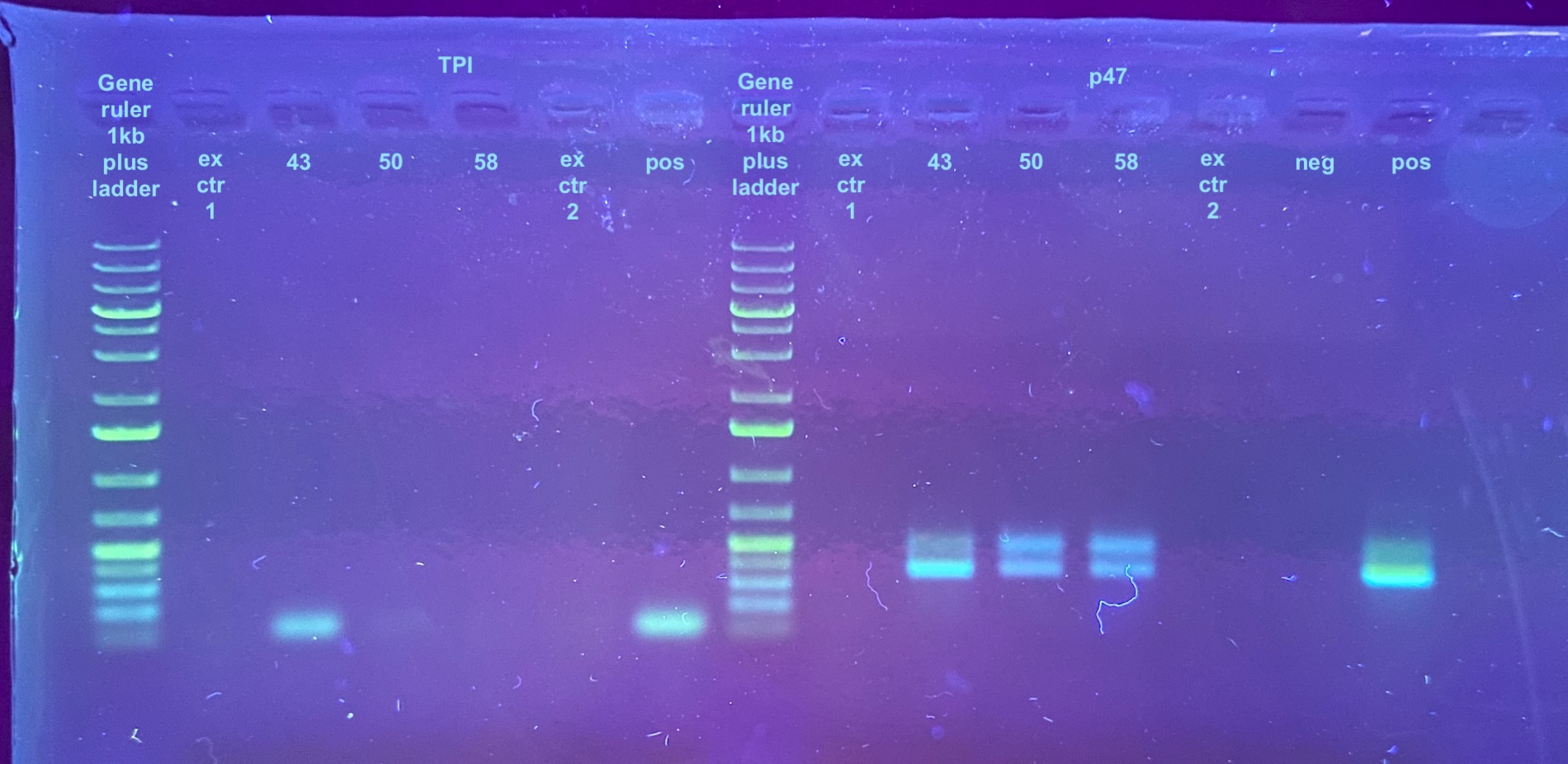
PCRs 20231130
Each sample was diluted 1:1 in water for PCR to conserve the sample. The positive control was not diluted. The PCR programs were set to 30 cycles, but these might need to be changed in the future because of the low DNA level. Information on the primers and their programs can be found here, but the TPI program had an extension time of 30 seconds.
The PCR process followed this general protocol. Afterwards a 1% gel was run for 35 minutes at 90V. I accidentally took the p47 samples out of the PCR machine while they were still in the extension time of the program, which is why I think there is a double band in these. I have never had an issue with double bands with this primer before, or with the positive control sample even, which makes me think it was because of the extension somehow and that the PCR should be considered normal.