DNA Extraction and PCRs of Samples from Day 0 and 21 Days from DiNV Experimental Evolution in Dinn Cells
Samples were taken at the 3 week mark and day 0 before adding virus. There are two replicate 50ul samples per flask, as well as the fly homogenate sample. For the DNA extractions, I chose 1 replicate from these samples as well as the fly homogenate to extract.
Samples:
| sample # | day | flask |
| 4 | NA | fly homogenate |
| 5 | day 0 | A |
| 7 | day 0 | B |
| 9 | day 0 | C |
| 11 | day 21 | A |
| 13 | day 21 | B |
| 15 | day 21 | C |
All samples were thawed on ice, this general protocol was used, and fliter tips were used at all times. Samples were ordered so that the virus infected samples were done second to the day 0 samples. 2 Extraction controls were done at the same time as well.
20231018 TPI and p47 PCRs
Because we might want to use the samples for a lot of PCRs and potentially DNA sequencing, Rob suggested I dilute the DNA 1:10 for the PCR. For every sample (including the extraction controls) I made a set of strip tubes with 18ul of DNA hydration solution. I put 2ul of DNA in each and mixed them. These were the samples I used for PCR.
I used sample 42 at 0.5ng dilution from the qPCR experiment as a positive control for TPI and p47. Water was the negative control.
The PCR process followed this general protocol.
TPI master mix
- 60ul GoTaq
- 3ul TPI F
- 3ul TPI R
- 42ul molecular grade water
p47 master mix
- 60ul GoTaq
- 3ul p47 F
- 3ul 47 R
- 42ul molecular grade water
All other steps followed the protocol. 1ul of the 1:10 diluted DNA was used for each sample. The positive control was not diluted. Each PCR program was ran for 30 cycles. Information on the primers and their programs can be found here
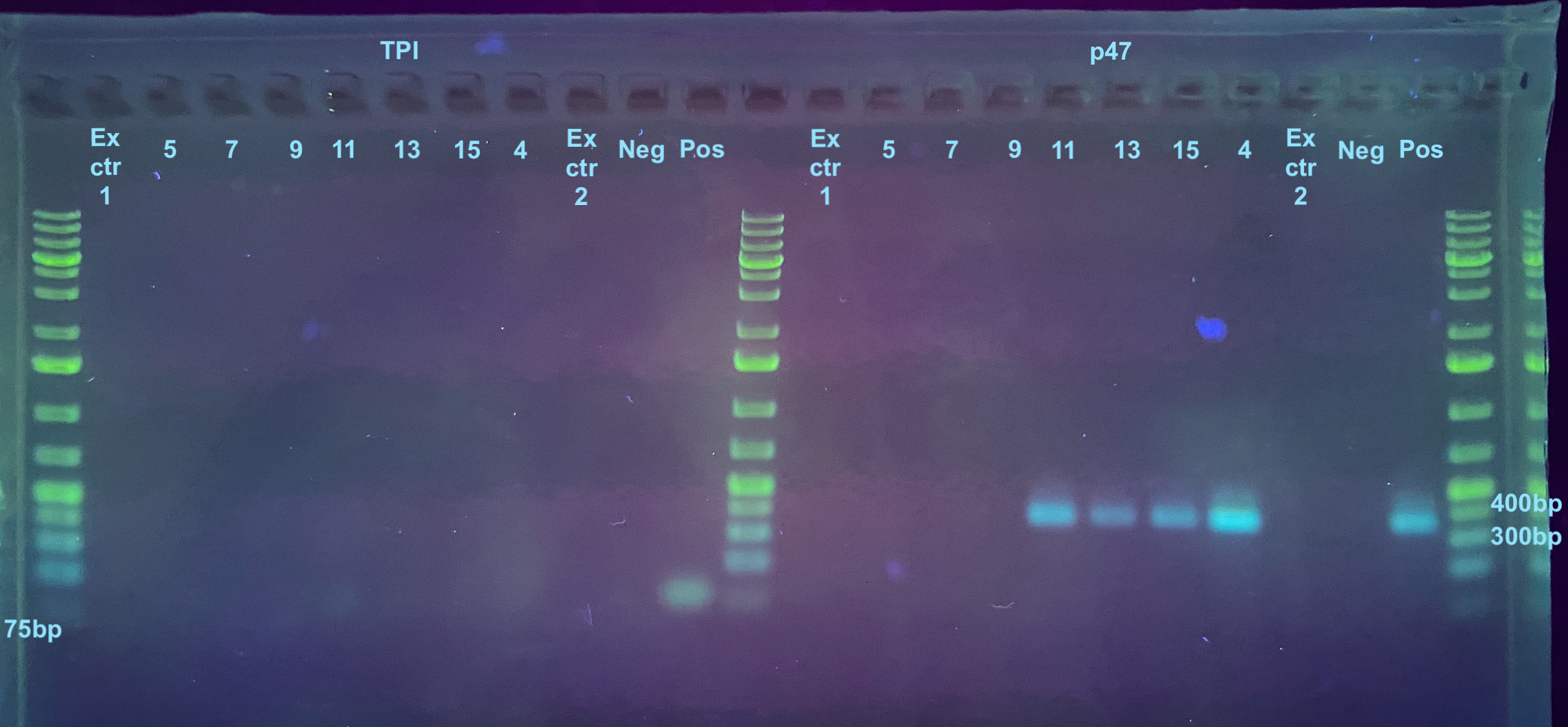
After the PCRs, a 1% small rectangle gel was made (30mL 1X TAE, 0.3g agarose, 0.75ul Midori stain) and run for 35 minutes at 90V.
This was not exactly what I expected. I think doing a 1:10 dilution on the DNA was diluting it too much. I will go back and PCR each one again with 35 cycles for both primers at a 1:2 dilution.
20231020 Redo TPI and p47 PCRs
I went back and diluted each DNA sample 1:2 (1ul of DNA and 1ul of DNA hydration solution). I did not dilute the positive control. I also changed the cycle numbers in both programs to 35. Information on the primers and their programs can be found here
The PCR process followed this general protocol.
TPI master mix
- 60ul GoTaq
- 3ul TPI F
- 3ul TPI R
- 42ul molecular grade water
p47 master mix
- 60ul GoTaq
- 3ul p47 F
- 3ul 47 R
- 42ul molecular grade water
After the PCRs, a 1% small rectangle gel was made (30mL 1X TAE, 0.3g agarose, 0.75ul Midori stain) and run for 35 minutes at 90V.