115, RP49, RPL11, and Lambda PCRs on Myd88 and Dv-1 Cell Samples from 12-Well Plate Infection Experiment
Using samples from this extraction from the Feb. 8th infection experiment.
20230220 115, RP49, and RPL11 PCRs
I think that the RP49 and Lambda primers can use the 115 program for PCR because their Tms are 60 and 62C, meaning a 55C annealing temp should work for them.
There are 48 samples to run for 115, and 24 for each RP49 and Lambda. I will have to do only 2 of these together because I will also need positive and negative controls for each.
I designed a 96 well plate layout for 115 and RP49:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1 | 9 | 17 | 25 | 33 | 41 | NEG | 2 | 18 | 34 | NEG | |
| B | 2 | 10 | 18 | 26 | 34 | 42 | POS | 4 | 20 | 36 | POS | |
| C | 3 | 11 | 19 | 27 | 35 | 43 | 6 | 22 | 38 | |||
| D | 4 | 12 | 20 | 28 | 36 | 44 | 8 | 24 | 40 | |||
| E | 5 | 13 | 21 | 29 | 37 | 45 | 10 | 26 | 42 | |||
| F | 6 | 14 | 22 | 30 | 38 | 46 | 12 | 28 | 44 | |||
| G | 7 | 15 | 23 | 31 | 39 | 47 | 14 | 30 | 46 | |||
| H | 8 | 16 | 24 | 32 | 40 | 48 | 16 | 32 | 48 | |||
| 115 | RP49 |
I used sample #8 from the previous experiment that had virus replication for the 115 positive control, along with another positive.
I used sample 16 from the same previous experiment for the RP49 control as a Myd88 sample.
All samples and reagents were thawed on ice, vortexed, and spun down before use.
All primers are diluted to 10uM.
- 115 master mix, n = 57
- 5ul GoTaq * 57 = 285ul
- 0.25ul 115 F * 57 = 14.25ul
- 0.25ul 115 R * 57 = 14.25ul
- 0.25ul nuclease free water * 57 = 199.5ul
- The master mix was vortexed, spun down, and kept on ice
- 9ul of the master mix went into the wells of the plate in the orientation as seen above
- 1ul of DNA was added to each well from the appropriate sample
- 1ul of nuclease free water was used as the negative control
- 1ul of the positive controls were used
- The wells were pipette mixed with a multichannel
- RP49 master mix, n= 29
- 5ul GoTaq * 29 = 145ul
- 0.25ul RP49 F * 29 = 7.25ul
- 0.25ul RP49 R * 29 = 7.25ul
- 3.5ul nuclease free water * 29 = 101.5
- The master mix was vortexed, spun down, and kept on ice
- 9ul of the master mix went into the wells of the plate in the orientation as seen above
- 1ul of DNA was added to each well from the appropriate sample
- 1ul of nuclease free water was used as the negative control
- 1ul of the positive controls were used
- The wells were pipette mixed with a multichannel
- The plate was sealed with a foil seal and placed in the thermocycler under the 115 program:
- 95 degrees C 5 minutes
- 95 degrees C 30 seconds
- 55 degrees C 30 seconds
- 72 degrees C 30 seconds
- 72 degrees C 5 minutes
- Hold at 12 degrees C
- italic lines are cycled 34 times
- Afterwards, I noticed that the top and bottom rows of the plate looked like they had evaporated, some a lot and some not so much. I made note so I could re-do those samples.
- RPL11 PCR
- Layout for strip tubes:
| Strip tube 1 | Strip tube 2 | Strip tube 3 | Strip tube 4 |
|---|---|---|---|
| 1 | 17 | 33 | NEG |
| 3 | 19 | 35 | POS |
| 5 | 21 | 37 | |
| 7 | 23 | 39 | |
| 9 | 25 | 41 | |
| 11 | 27 | 43 | |
| 13 | 29 | 45 | |
| 15 | 31 | 47 |
- RPL11 master mix, n= 29
- 5ul GoTaq * 29 = 145ul
- 0.25ul RPL11 F * 29 = 7.25ul
- 0.25ul RPL11 R * 29 = 7.25ul
- 3.5ul nuclease free water * 29 = 101.5
- The master mix was vortexed, spun down, and kept on ice
- 9ul of the master mix went into the wells of the plate in the orientation as seen above
- I used sample 8 from the previous experiment for this positive control
- 1ul of DNA was added to each well from the appropriate sample
- 1ul of nuclease free water was used as the negative control
- 1ul of the positive controls were used
- The wells were vortex mixed and spun down
- The tubes were placed in the thermocycler under the RPL11 program:
- 95 degrees C 5 minutes
- 95 degrees C 30 seconds
- 57 degrees C 1 minute
- 72 degrees C 30 seconds
- 72 degrees C 5 minutes
- Hold at 12 degrees C
- italic lines are cycled 34 times
- Samples were placed in the fridge overnight
20230221 Lambda and Redo PCRs
- Lambda PCR
- Layout for strip tubes:
| Strip tube 1 | Strip tube 2 | Strip tube 3 | Strip tube 4 |
|---|---|---|---|
| 1 | 9 | 29 | NEG |
| 2 | 10 | 30 | POS |
| 3 | 11 | 31 | |
| 4 | 12 | 32 | |
| 5 | 25 | 33 | |
| 6 | 26 | 34 | |
| 7 | 27 | 35 | |
| 8 | 28 | 36 |
- Lambda master mix, n= 29
- 5ul GoTaq * 29 = 145ul
- 0.25ul Lambda F * 29 = 7.25ul
- 0.25ul Lambda R * 29 = 7.25ul
- 3.5ul nuclease free water * 29 = 101.5
- The master mix was vortexed, spun down, and kept on ice
- 9ul of the master mix went into the wells of the plate in the orientation as seen above
- I used sample 7 from the previous experiment for this positive control
- 1ul of DNA was added to each well from the appropriate sample
- 1ul of nuclease free water was used as the negative control
- 1ul of the positive controls were used
- The wells were vortex mixed and spun down
- The tubes were placed in the thermocycler under the 115 program as shown above
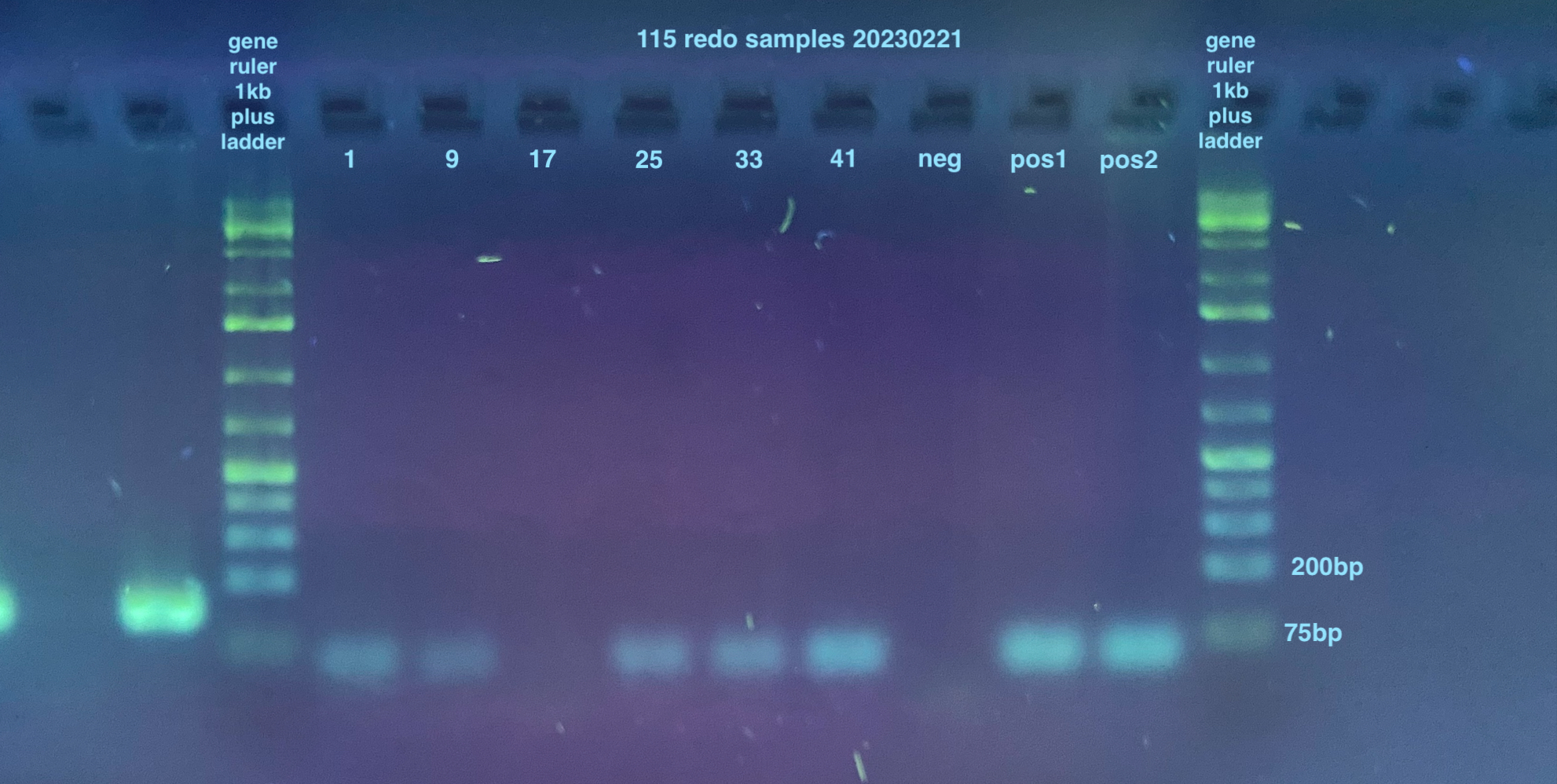
- Redo 115 PCR:
- Samples were the ones from the top row of the plate from the day before that had evaporated, so: 1, 9, 17, 25, 33, 41, and a negative and positive. I had forgotten to do the bottom of the plate samples, but because I will be running these on qPCR for a more accurate answer, I ended up not running those (also they were all non-infected so they should not amplify)
- 115 master mix, n= 9
- 5ul GoTaq * 9 = 45ul
- 0.25ul 115F * 9 = 2.25ul
- 0.25ul 115 R * 9 = 2.25ul
- 3.5ul nuclease free water * 9 = 31.5ul
- The master mix was vortexed, spun down, and kept on ice
- 9ul of the master mix went into the wells of the plate in the orientation as seen above
- I used the 3mL positive control sample
- 1ul of DNA was added to each well from the appropriate sample
- 1ul of nuclease free water was used as the negative control
- 1ul of the positive controls were used
- The wells were vortex mixed and spun down
- The tubes were placed in the thermocycler under the 115 program as shown above
- Redo RP49 PCR:
- Samples were the ones from the top row of the plate from the day before that had evaporated, so: 2, 18, 34, 38, and a negative and positive. I had forgotten to do the bottom of the plate samples, but because I will be running these on qPCR for a more accurate answer, I ended up not running those
- RP49 master mix, n= 6
- 5ul GoTaq * 6 = 30ul
- 0.25ul RP49 F * 6 = 1.5ul
- 0.25ul RP49 R * 6 = 1.5ul
- 3.5ul nuclease free water * 9 = 21ul
- The master mix was vortexed, spun down, and kept on ice
- 9ul of the master mix went into the wells of the plate in the orientation as seen above
- I used sample 16 from the previous experiment as the positive control
- 1ul of DNA was added to each well from the appropriate sample
- 1ul of nuclease free water was used as the negative control
- 1ul of the positive controls were used
- The wells were vortex mixed and spun down
- The tubes were placed in the thermocycler under the 115 program as shown above
20230221 Gels
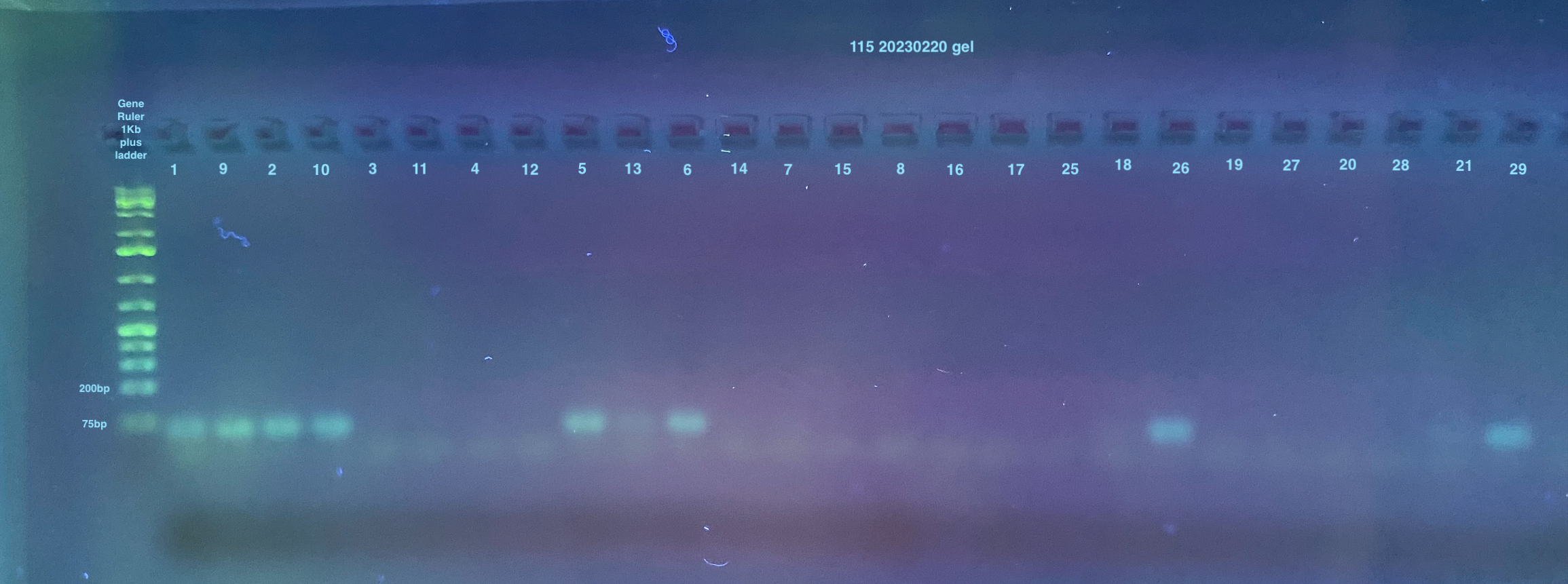
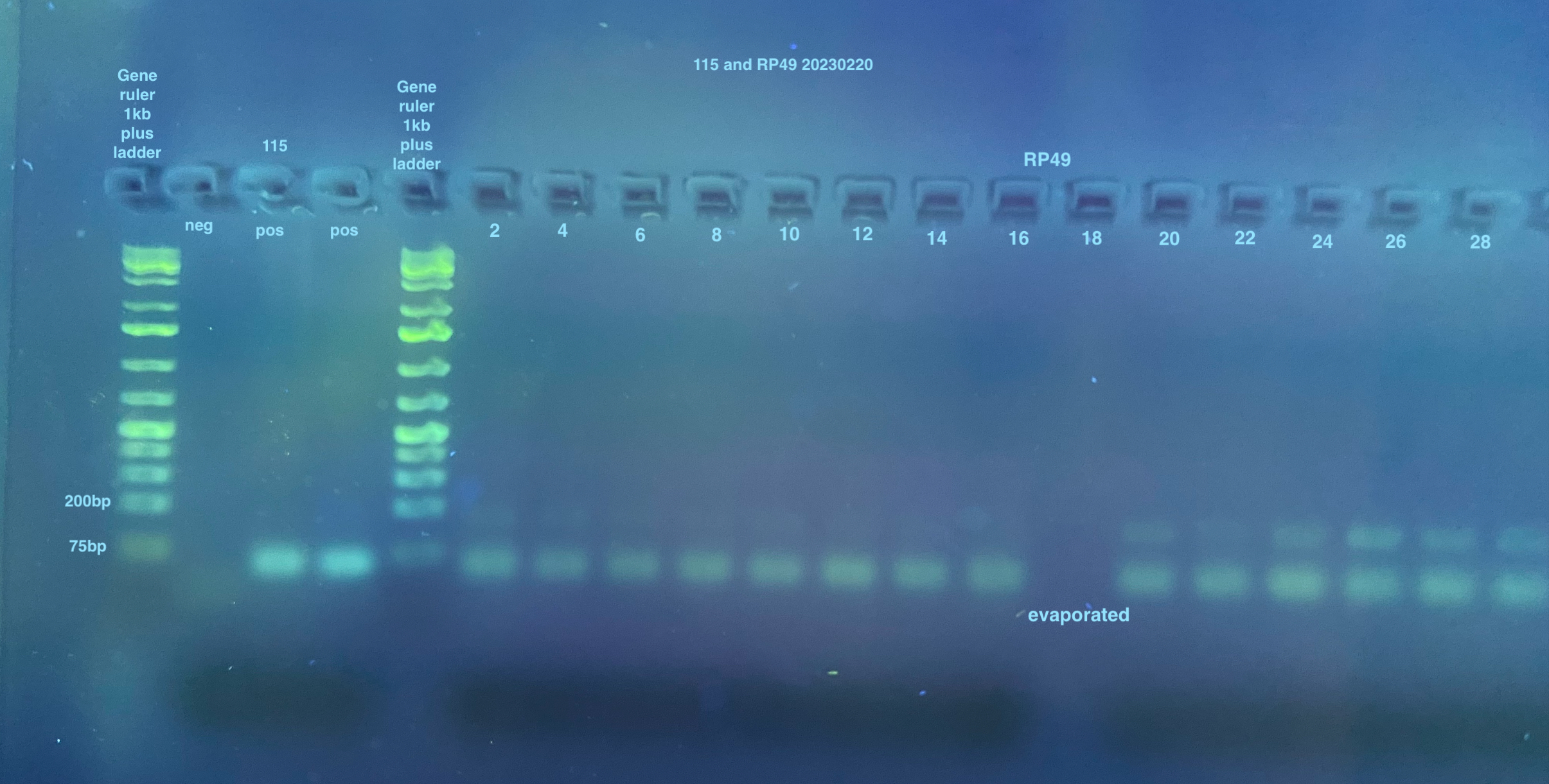
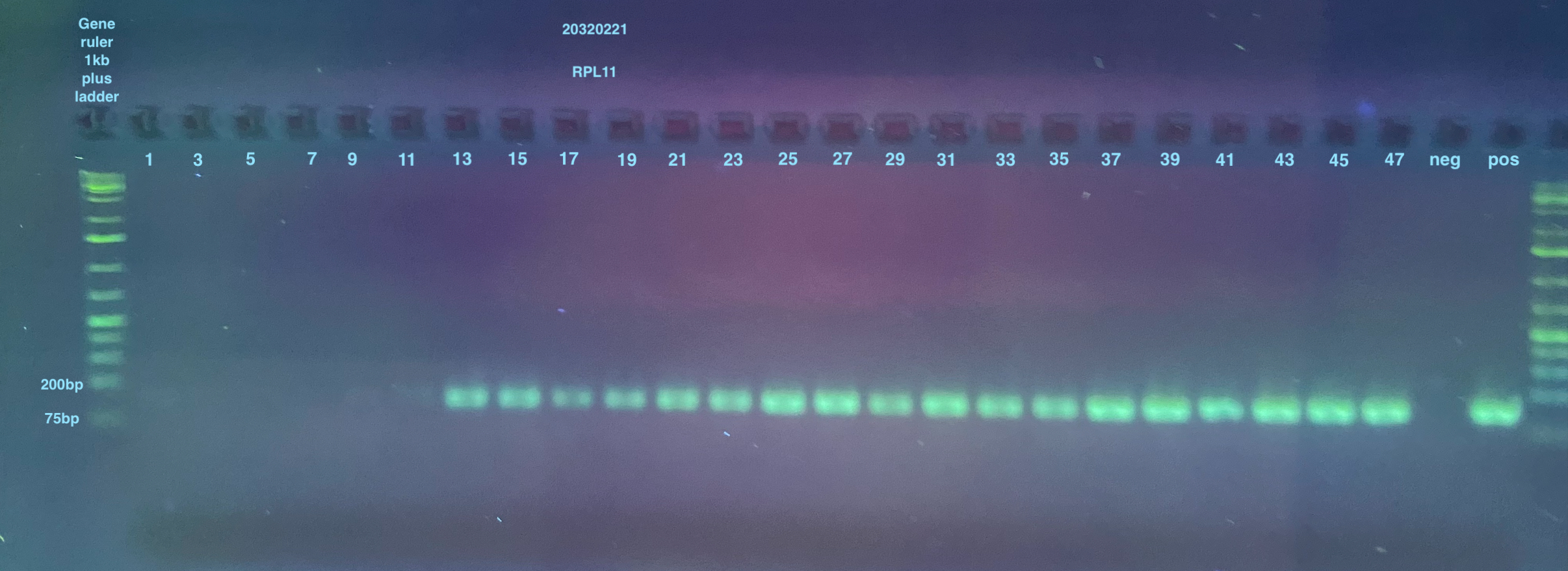
2% large gels were run for all samples, I used 2 of them. Each used 160mL of 1X TAE, 3.2g of agarose, and 4ul of Midori stain.
The first gel has some strange ordering of samples because I used the multichannel that doesn’t work very well and some of the evaporated samples didn’t pipette at all into the wells (no liquid left)
Gel of 115 and RP49 from 20230220 PCRS:


Gel of RPL11, Lambda, and RPL49 and 115 Redos:
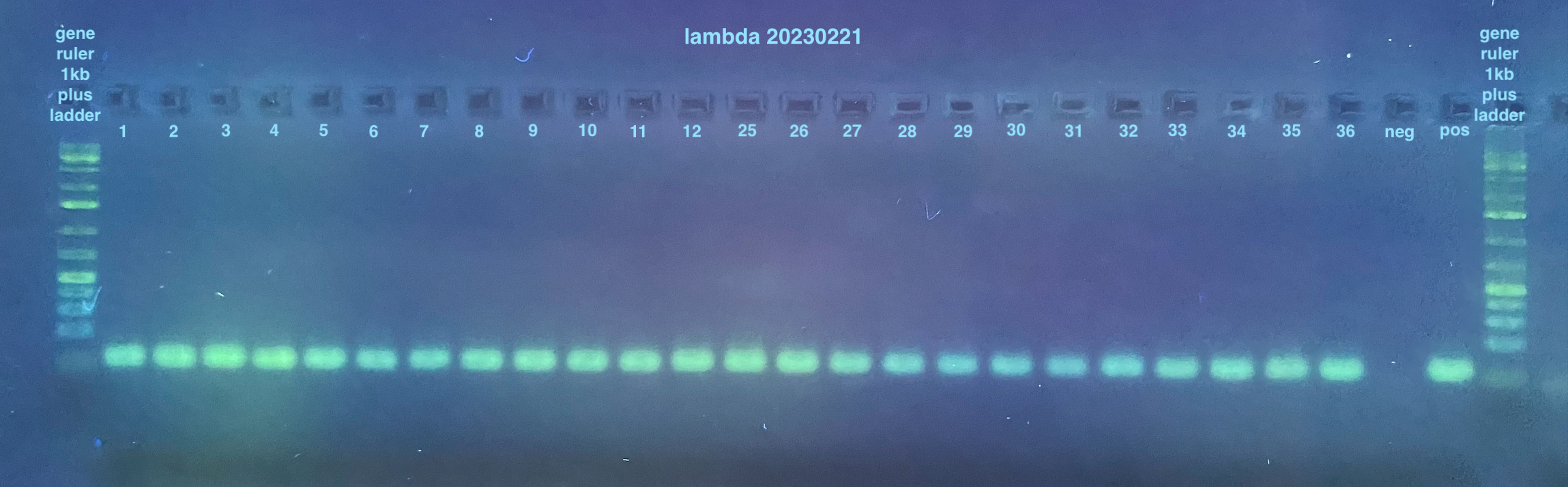
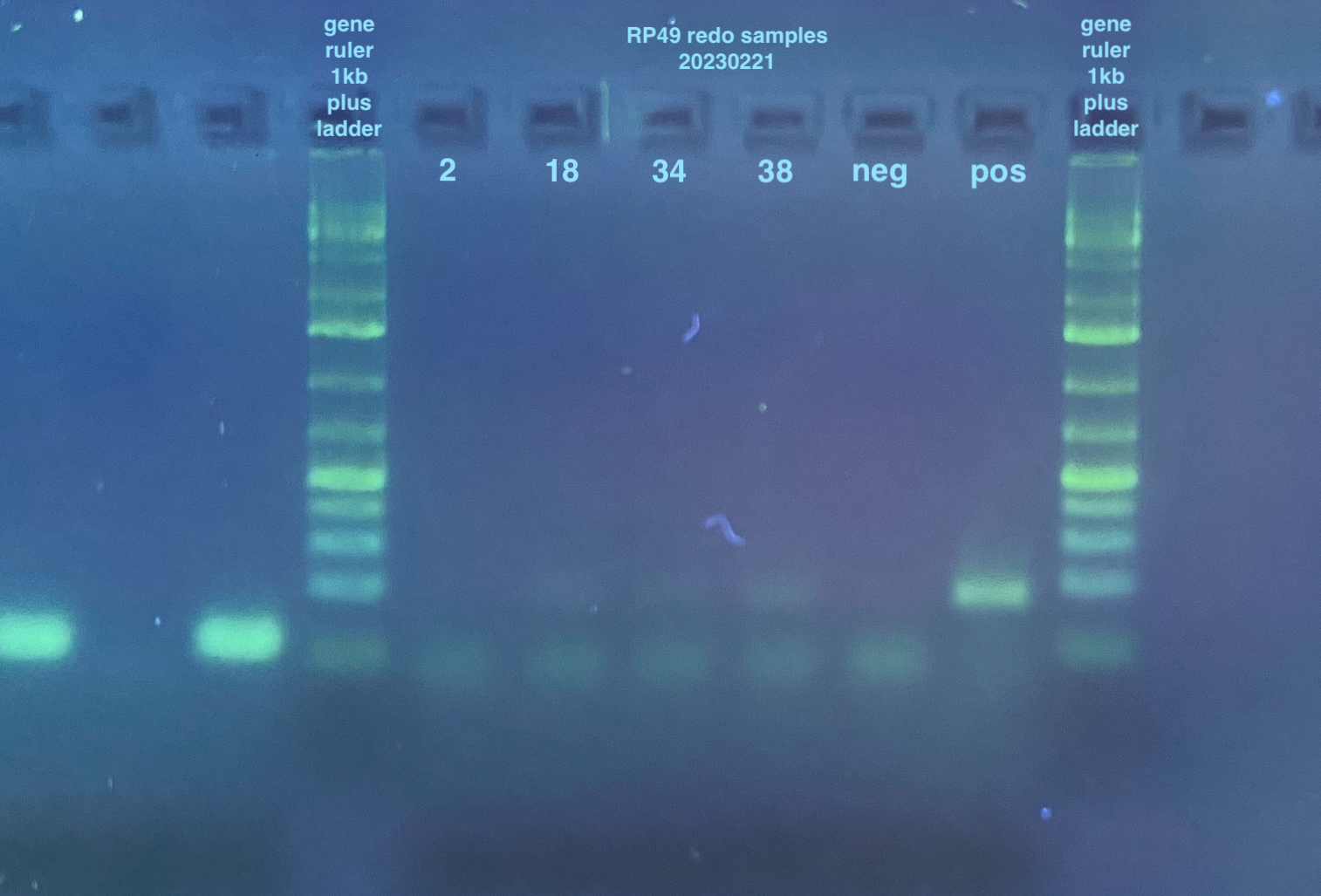
I scored the results from these gels as either yes, no, or maybe for a band and put the data into this spreadsheet. I then calculated percentages of yes/no/maybe for each “treatment” by collapsing the replicates (three per treatment) so it is either 0%, 33%, 66%, or 100% for yes/no/maybe. Those results are stored here and the visualized results in graphs can be seen here.