Doing qPCR on DNA Extracts of Dv-1 and Myd88 Cells, Infected with DiNV and Control
Because of the variability between the extraction replicates, and the number of PCRs I need to do, I am not using the extraction replicates for this process. Just one DNA sample per sample.
Sample inforation and plate planning is here and here
Diluting Samples
- Want to have all samples at 1ng/ul to use for input
- For the samples that are too low, I did not do anything with them and used them as-is for the qPCR
- All samples were thawed on ice, vortexed and spun down before use
- I made new dilution tubes for the samples needing a dilution, and used the DNA hydration solution from the Puregene DNA extraction kit to do the elution
| Sample_ID | qubit_quantity | ul_EB_to_1ng/ul | ul_DNA |
|---|---|---|---|
| 1 | too low | NA | NA |
| 2 | too low | NA | NA |
| 3 | too low | NA | NA |
| 4 | 5.16 | 15.48 | 3 |
| 5 | too low | NA | NA |
| 6 | 48.1 | 96.2 | 2 |
| 7 | 13.2 | 26.4 | 2 |
| 8 | 44.3 | 88.6 | 2 |
| 9 | 10.6 | 21.2 | 2 |
| 10 | 45.6 | 91.2 | 2 |
| 11 | 11.2 | 22.4 | 2 |
| 12 | 20.1 | 40.2 | 2 |
| 13 | 14.5 | 29 | 2 |
| 14 | 18.3 | 36.6 | 2 |
| 15 | 14.2 | 28.4 | 2 |
| 16 | 14 | 28 | 2 |
- All samples were kept on ice
qPCR Plates
- For all samples:
- 3 replicate reactions per PCR primer
- Need to have 115 (DiNV) reactions
- Need to have a cell-line specific reaction (either RP49 for Myd88 or RPL11 for Dv-1)
- For supernatant samples only, need to have Lambda reaction
- Because of the number of samples, this was spit up over 2 plates and were laid out like this:

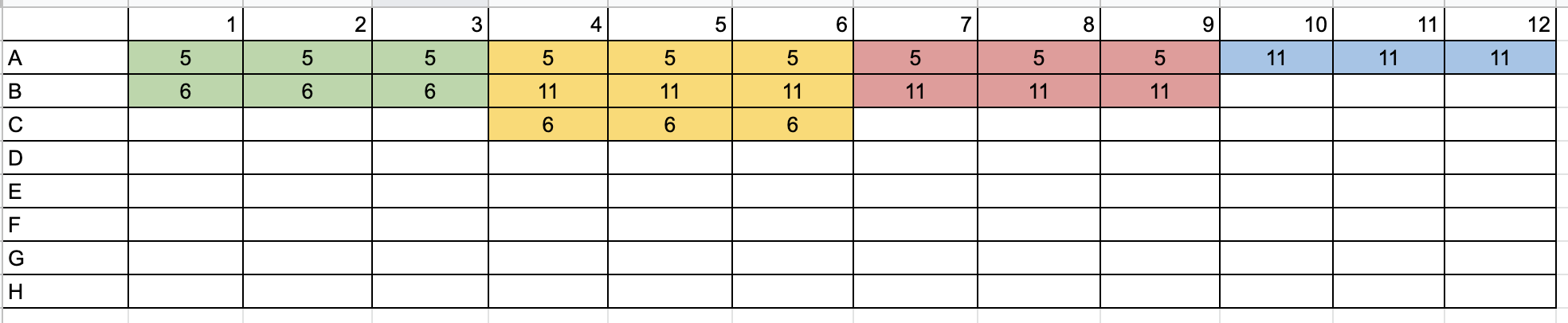
Master Mixes Plate 1
- I made 10uM aliquots of the Lambda (from Kent’s box) and RP49 ( number 8 and 9 on lab spreadsheet) primers
- 10ul of 100uM stock
- 90ul nuclease free water
- Made master mixes for each plate separately (done a few hours apart)
- All components were vortexed and spun down before use
- Sample numbers for plate 1:
- Lambda is 18 + 2 for error: 20
- RPL11 is 18 + 2 for error: 20
- 115 is 39 + 4 for error: 43
- RP49 is 21 + 2 for error: 23
- 115 master mix:
- 5ul Sso (supermix) * 43 = 215ul
- 0.5ul 115 F primer * 43 = 21.5ul
- 0.5ul 115 R primer * 43 = 21.5ul
- 3ul nuclease free water * 43 = 129ul
- RP49 master mix:
- 5ul Sso (supermix) * 23 = 115ul
- 0.5ul RP49 F primer * 23 = 11.5ul
- 0.5ul Rp49 R primer * 23 = 11.5ul
- 3ul nuclease free water * 23 = 69ul
- RPL11 master mix:
- 5ul Sso (supermix) * 20 = 100ul
- 0.5ul RPL11 F primer * 20 = 10ul
- 0.5ul RPL11 R primer * 20 = 10ul
- 3ul nuclease free water * 20 = 60ul
- Lambda master mix:
- 5ul Sso (supermix) * 43 = 100ul
- 0.5ul Lambda F primer * 43 = 10ul
- 0.5ul Lmbda R primer * 43 = 10ul
- 3ul nuclease free water * 43 = 60ul
- Vortexed and spun down mixes and kept on ice
Plate set up
- Used specific qPCR plate and seal
- Put 9ul of primer mix into correct wells (see plate picture above)
- Added 1ul of sample to each well (see plate picture above)
- Pipette mixed each well with 5ul using a multichannel
- Spun down the plate for 1 min at 4000g
At the qPCR machine
- Logged into computer
- Clicked on the BioRAD CFX manager software
- Chose user-defined protocol, existing protocol, and KMM folders
- Selected the p47 program
- Used the button on the lid to open the machine
- Placed the plate in correct orientation
- Pressed the button on the machine to close it
- Started the program on the computer
- Ran for 1.5ish hours, and saved the results on my folder on the computer and put on a flash drive
Master Mixes Plate 2
- All components were vortexed and spun down before use again and kept on ice the whole time
- Sample numbers for plate 2:
- Lambda is 6 + 1 for error: 7
- RPL11 is 6 + 1 for error: 7
- 115 is 9 + 1 for error: 10
- RP49 is 3 + 1 for error: 4
- 115 master mix:
- 5ul Sso (supermix) * 10 = 50ul
- 0.5ul 115 F primer * 10 = 5ul
- 0.5ul 115 R primer * 10 = 5ul
- 3ul nuclease free water * 10 = 30ul
- RP49 master mix:
- 5ul Sso (supermix) * 4 = 20ul
- 0.5ul RP49 F primer * 4 = 2ul
- 0.5ul Rp49 R primer * 4 = 2ul
- 3ul nuclease free water * 4 = 12ul
- RPL11 master mix:
- 5ul Sso (supermix) * 7 = 35ul
- 0.5ul RPL11 F primer * 7 = 3.5ul
- 0.5ul RPL11 R primer * 7 = 3.5ul
- 3ul nuclease free water * 7 = 21ul
- Lambda master mix:
- 5ul Sso (supermix) * 7 = 35ul
- 0.5ul Lambda F primer * 7 = 3.5ul
- 0.5ul Lmbda R primer * 7 = 3.5ul
- 3ul nuclease free water * 7 = 21ul
- Vortexed and spun down mixes and kept on ice
Plate set up
- Used specific qPCR plate and seal
- Put 9ul of primer mix into correct wells (see plate picture above)
- Added 1ul of sample to each well (see plate picture above)
- Pipette mixed each well with 5ul using a multichannel
- Spun down the plate for 1 min at 4000g
At the qPCR machine
- Logged into computer
- Clicked on the BioRAD CFX manager software
- Chose user-defined protocol, existing protocol, and KMM folders
- Selected the p47 program
- Used the button on the lid to open the machine
- Placed the plate in correct orientation
- Pressed the button on the machine to close it
- Started the program on the computer
- Ran for 1.5ish hours, and saved the results on my folder on the computer and put on a flash drive