Second Test of the Hirt Method for Viral DNA Extraction With Phenol Chloroform Purification on DV-2 Cells Infected with DiNV
Protocol modified from previous attempt with: 1. making sure to add 2X 100% ethanol during precipitation step 2. keeping the phenol chloroform cold 3. using phase lock tubes durning phase separation 4. mixing the tubes with the phenol/chloroform for 10 minutes with rotating mixing 5. increasing some centrifugation times 6. all centrifugations were at 4 degrees C except for the phase separation.
Chromosomal DNA Precipitation
- Placed the small centrifuge in the 4 degree fridge to cool it down
- Thawed 2 samples from the -80: DV-1 samples infected with DiNV from Kent
- 1.5mL sample: DiNV GS 20181123 Dv-1-2 day 10 lysate
- 0.7mL sample: DiNV GS 20181123 Dv-1-1 day 10 lysate
- Transferred the volume of the samples to 1.5mL tubes so they fit in the centrifuges. I used clipped pipette tips to protect long DNA pieces. Tubes are named by the volume of sample in the original sample tubes (roughly)
- Centrifuged tubes for 3 minutes at 5000rcf
- This may have been too hard of a centrifuge

- This may have been too hard of a centrifuge
- Removed the supernatant from the pellet
- Added 1.5mL of 1X PBS to the tubes to wash the cells
- Centrifuged tubes 3 minutes at 5000rcf
- Removed supernatant from the cell pellet
- Resuspended pellet in 147ul of 50mM Tris HCl, 10mM EDTA and 3ul of 5mg/mL RNase A (RNase A at a final concentration of 100ug/mL)
- Added equal volume (150ul) of 1.2% SDS to the tubes
- Inverted tubes to mix several time and spun down
- The cell pellets were a little stuck and I had to vortex them
- Incubated tubes on the bench for 5 minutes to lyse the cells
- Tubes “went clear” very quickly, the cells disappeared
- Precipitated chromosomal DNA and cellular debris by added 210ul of 3M CsCl, 1M potassium acetate, 0.67M acetic acid
- A white precipitate immediately started forming in the tubes

- A white precipitate immediately started forming in the tubes
- Inverted tubes briefly
- Immediately placed tubes on ice for 30 minute incubation (this was increased from 15 minutes)
- Spun down phase lock tubes at 12,000rpm for 30 seconds at room temp
- Centrifuged tubes for 30 minutes at 16,000rcf at 4 degrees C (this was increased from 15 minutes and the spin was now at 4 degree)
- Removed supernatant with clipped pipette tips to phase lock tubes (~500ul)
Phenol Chloroform Extraction and DNA Precipitation
- Took tubes and setup to the hood
- Added equal volume (500ul) of cold phenol-chlorofomr-isoamy-alcohol to the tubes
- Mixed tubes up and down and then put them on an orbital mixer in the hood for 10 minutes
- Opened caps after mixing to release gas
- Centrifuged tubes at 16,000rcf at room temp for 15 minutes
- Looked for phase separation
- This was a little strange. There was phase separation in both tubes (good) but I don’t think the phase lock tubes worked properly. All the gel was on the top of the liquid, and I’m pretty sure there is also supposed to be a layer of gel between the two phases to protect from taking the bottom one. I began to wonder if the chemistry of the solutions was somehow not right
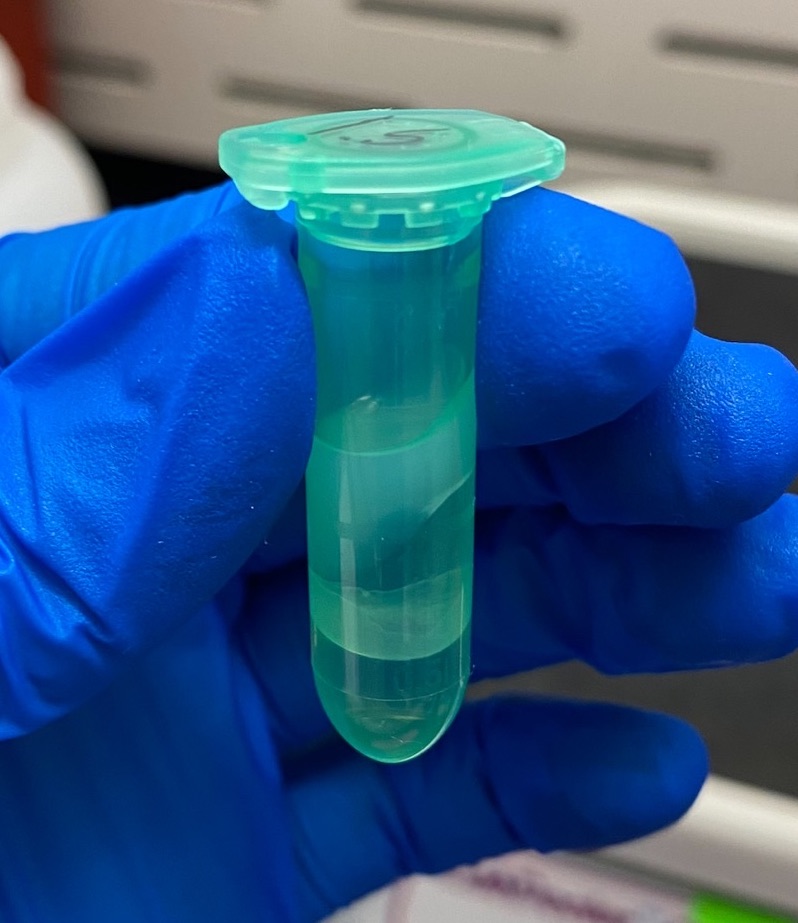

- This was a little strange. There was phase separation in both tubes (good) but I don’t think the phase lock tubes worked properly. All the gel was on the top of the liquid, and I’m pretty sure there is also supposed to be a layer of gel between the two phases to protect from taking the bottom one. I began to wonder if the chemistry of the solutions was somehow not right
- Transferred as much liquid as possible from the top phase to new 1.5mL. I had to sweep away the gel layer on top first, this was a little hard but not unmanageable. I also forgot to use clipped pipette tips. This was about 400-450ul
- Added 0.1X (40 or 45ul) of 3M NaOAc to each tube
- Added ~900ul of cold 100% ethanol to each tube
- Gently inverted to mix tubes
- Stored tubes in the -80 for 2 hours
- These tubes froze again, I’m not sure if they were supposed to or not
- Centrifuged tubes at 4 degrees C for 20 minutes at 16,300rcf
- Tubes had huge white pellet after this, larger than the cell pellets so this is definitely not all DNA
- Pipetted off supernatant
- Added 500ul of fresh cold 80% ethanol
- Centrifuged tubes 30 min, 16,300rcf at 4 degrees C
- Removed all ethanol
- Let tubes dry ~30 minutes
- Resuspended in 75ul of 10mM tris HCl and let sit overnight
- Qubit the next day:
- 1.5mL sample: 16.3ng/ul
- 0.7mL sample: 11.5ng/ul
- Samples are stored in the fridge
20220909 p47 and RPL11 PCRs
- p47
- So I have 2 samples, plus 1 for a positive control, and plus 1 for a negative control
- Made master mix on ice:
- 5ul GoTaq * 4.5 = 22.5ul
- 0.25ul p47_F * 4.5 = 1.125ul
- 0.25ul p47_R * 4.5 = 1.125ul
- 3.5ul molecular grade water * 4.5 = 15.75ul
- Vortexed and spun down master mix
- Added 9ul master mix to 4 strip tubes
- Added 1ul of DNA sample to appropriate tubes
- Added 1ul of 3mL HMW extraction to the positive control tube
- Added 1ul of molecular grade water to the negative control tube
- Vortexed and spun down strip tubes
- Placed tubes in the p47 PCR program
- RPL11
- Using the same sample scheme as above
- Made master mix on ice:
- 5ul GoTaq * 4.5 = 22.5ul
- 0.25ul p47_F * 4.5 = 1.125ul
- 0.25ul p47_R * 4.5 = 1.125ul
- 3.5ul molecular grade water * 4.5 = 15.75ul
- Vortexed and spun down master mix
- Added 9ul master mix to 4 strip tubes
- Added 1ul of DNA sample to appropriate tubes
- Added 1ul of 3mL HMW extraction to the positive control tube
- Added 1ul of molecular grade water to the negative control tube
- Vortexed and spun down strip tubes
- Placed tubes in the RPL11 PCR program
- Ran a 2% gel for 35 minutes at 90 volts after the PCR programs were done:
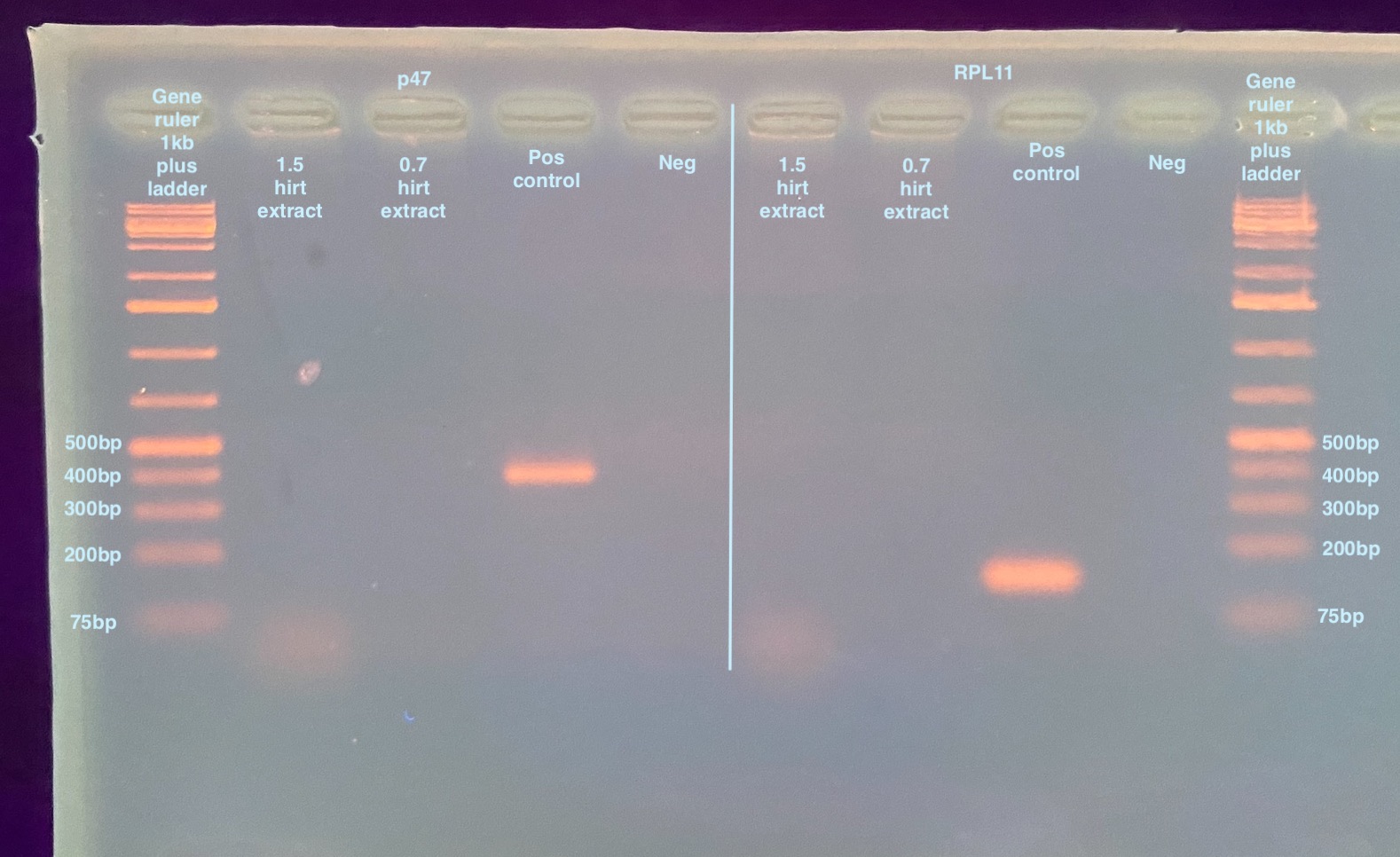
Neither sample amplify any DNA. I don’t think this is because no DNA is in there, I think that whatever precipitates out with the DNA is inhibiting the PCR reaction. I can try to dilute the DNA solution to see if that ameliorates the issue. The same thing happened in the previous extraction PCRs where the sample that had the huge pellet after ethanol precipitation also did not amplify for either primers. I think though that my main goal should be to try to remove all those precipitates prior to doing the final precipitation, and trying to optimize to remove all the chromosomal DNA. I think that the ice incubation and centrifugation after adding the 3M CsCl, 1M potassium acetate, 0.67M acetic acid is enough to get both the liquid and the chromosomal DNA out of the solution and I think that is precipitating when I’m trying to just get the DNA.