Testing the Hirt Method for Viral DNA Extraction With Phenol Chloroform Purification on DV-2 Cells Infected with DiNV
Protocol is one I came up with by looking at A, B, C, and D. Clearly some things did not work right.
Preparing Solutions 20220718
Need 50mM Tris HCl pH 7.5, 10mM EDTA for lysis
Making 10mL solution:
- 500ul of 1M Tris
- 200ul of 0.5M EDTA
- 9.3mL of molecular grade water to 10mL
Need 1.2% SDS for lysis, already have 10% solution
Making 50mL solution:
- 6mL of 10% solution
- 44mL of molecular grade water
Need 3M CsCl, 1M potassium acetate, 0.67 acetic acid for chromosomal DNA precipitation
There are no stock solutions of CsCl or potassium acetate so first I’ll have to make those
5M CsCl solution in 10mL:
- Molecular mass is 186.36g/mol
- So that’s 186.36 grams in 1 liter for a 1M solution
- A 5M solution in 1 liter is 186.36g * 5 = 931.8g
- Now divide by 100 to get the grams for 10mL
- 931.8g CsCl / 100 = 9.318 grams CsCl
- And I’ll need molecular grade water up to 10mL
5M potassium acetate in 10mL:
- Molecular mass is 98.15g/mol
- 5M solution in 1 liter is 98.15g * 5 = 490.75g
- Divide by 100: 490.75g / 100 = 4.91g potassium acetate
- And molecular grade water up to 10mL
Make 10mL of 3M CsCl, 1M potassium acetate, 0.67M acetic acid:
- 6mL of 5M CsCl
- 2mL of 5M potassium acetate
- 0.465mL of glacial (17.4M) acetic acid
- 1.18mL of molecular grade water
All solutions were prepared in the hood just in case some reaction happened.
Test Extraction 20220720
Chromosomal DNA Precipitation
- Placed the small centrifuge in the 4 degree fridge to cool it down
- Thawed 2 samples from the -80: DV-1 samples infected with DiNV from Kent
- 1.2mL sample: DiNV GS 20181123 DV-1-3 day 10 lysate
- 0.2mL sample: DiNV GS 2.1 FH, 240hr flask 7-18-2018
- Transferred the volume of the samples to 1.5mL tubes so they fit in the centrifuges. I used clipped pipette tips to protect long DNA pieces. Tubes are named by the volume of sample in the original sample tubes (roughly)
- Centrifuged tubes for 4 minutes at 300rpm
- The cells didn’t really pellet well here, so I did a secondary centrifuge for 2 minutes at 500rpm
- Removed the supernatant from the pellet
- Here it was obvious that the pellet still wasn’t very good and I lost some cells
- Added 1.5mL of 1X PBS to the tubes to wash the cells
- Centrifuged tubes 4 minutes at 300rpm
- Again this was not long enough of a centrifuge, and I didn’t want to loose any more cells so I centrifuged it for 2 minutes at 5000rpm
- Removed supernatant from the cell pellet
- Resuspended pellet in 147ul of 50mM Tris HCl, 10mM EDTA and 3ul of 5mg/mL RNase A (RNase A at a final concentration of 100ug/mL)
- Added equal volume (150ul) of 1.2% SDS to the tubes
- Inverted tubes to mix several time and spun down
- Incubated tubes on the bench for 5 minutes to lyse the cells
- Tubes “went clear” very quickly, the cells disappeared
- The 1.2mL sample tube became very viscous
- Precipitated chromosomal DNA and cellular debris by added 210ul of 3M CsCl, 1M potassium acetate, 0.67 acetic acid
- A white precipitate immediately started forming in the tubes
- Inverted tubes briefly
- Immediately placed tubes on ice for 15 minute incubation
- Centrifuged tubes for 15 minutes at 16,000rcf
- Removed supernatant with clipped pipette tips to new 1.5mL tubes (~500ul)
Phenol Chloroform Extraction and DNA Precipitation
- Took tubes and setup to the hood
- Added equal volume (500ul) of phenol-chlorofomr-isoamy-alcohol to the tubes
- Vortexed the tubes gently for ~30 seconds
- Centrifuged tubes at maximum speed for 5 minutes
- Afterwards, only the 1.2mL tube had separated into phases
- 0.2mL sample
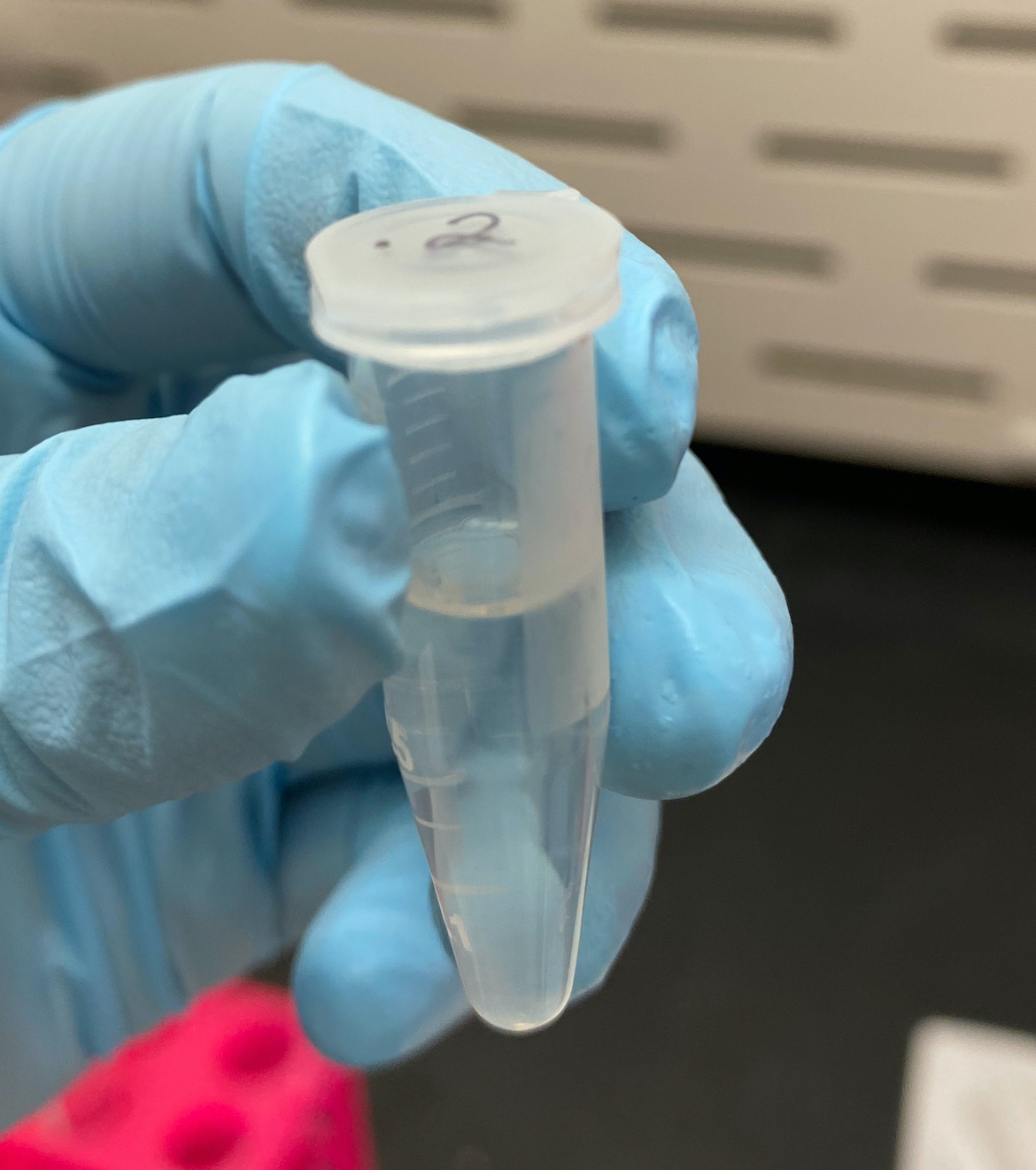
- 1.2mL sample

- Transferred as much liquid as possible from the top phase to new 1.5mL tubes using clipped pipette tips
- This was about 500ul for the 1.2mL tube, although it was hard to not mix the phases
- I did not know what to do for the .2 sample so I just took 500ul from the tube
- Added 0.1X (50ul) of 3M NaOAc to each tube
- Inverted to mix tubes
- Placed tubes in the -80 for 2 hours (note I was supposed to add ethanol before this step)
- Afterwards, both tubes were completely frozen (this should have been a red flag that I did something wrong!)
- Centrifuged tubes in 4 degree centrifuge for 30 minutes at 16,300rcf (max for that centrifuge)
- After spin:
- Things seemed really wrong, there was no precipitate (DNA) in either tube
- The 1.2mL tube had separated again into 2 layers!
- At first I assumed I did not add enough NaOAc for the DNA to precipitate, and I wondered if I did not separate out the two phases well enough in the one that separated again
- I decided to split up the top and bottom of the 1.2mL tube into separate tubes, and add 0.1X NaOAc to each tube again
- 0.2mL tube after NaOAc spin:
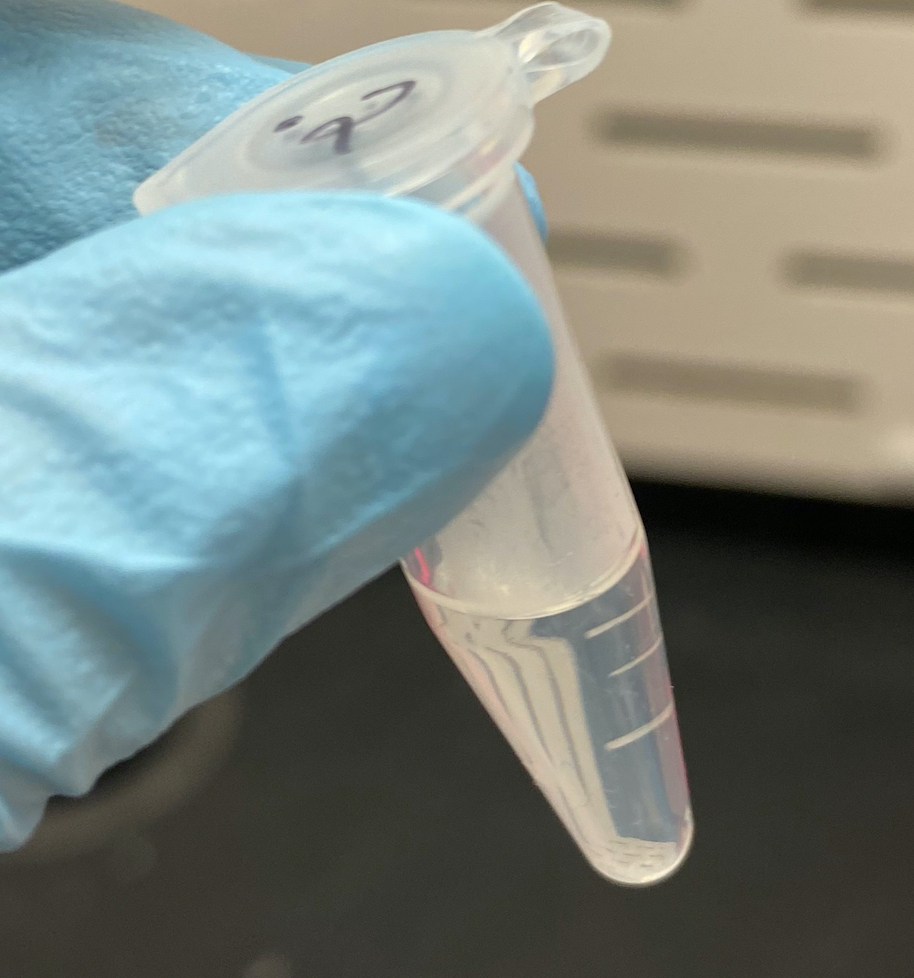
- 1.2mL tube after NaOAc spin
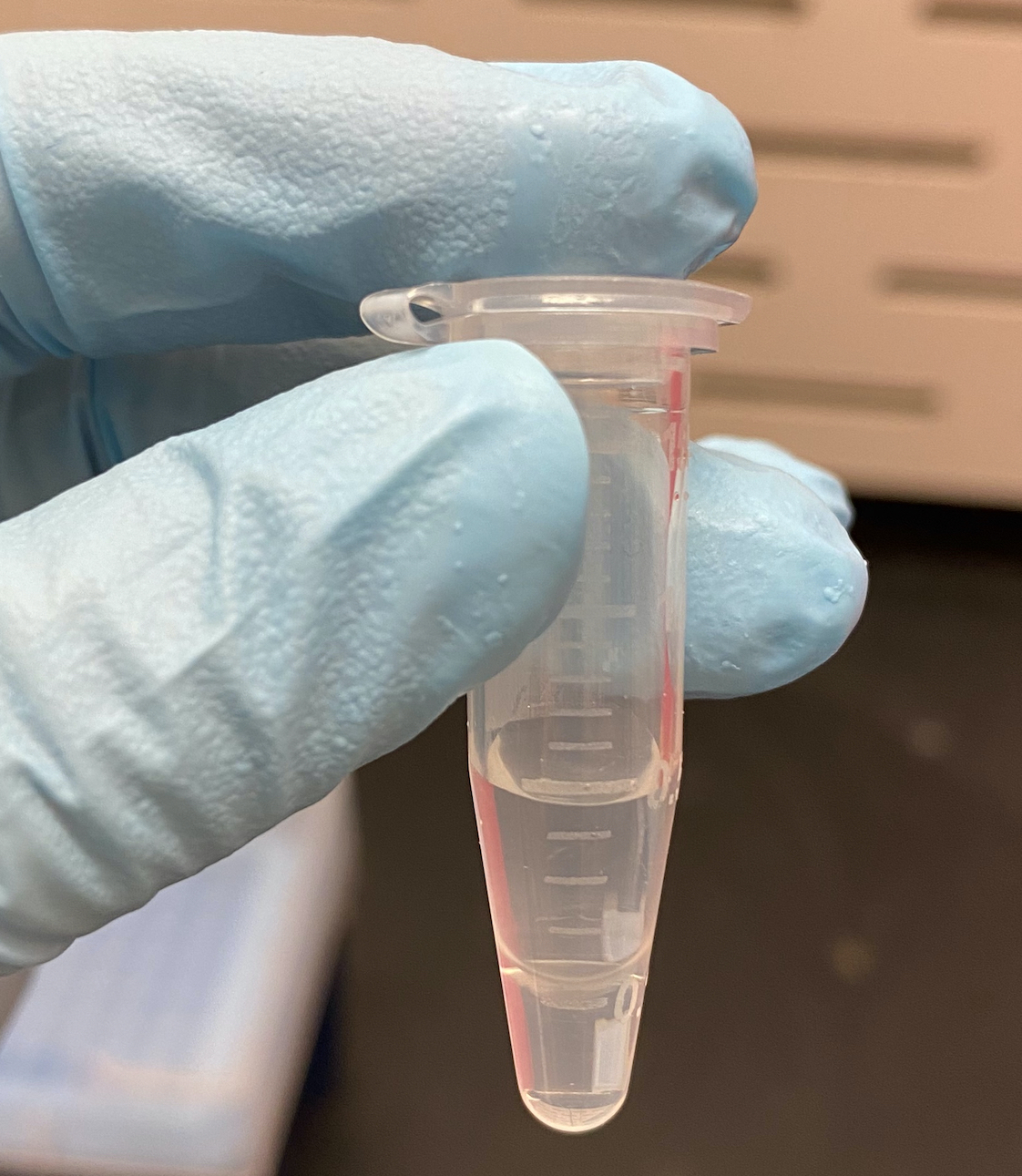
- I added another 50ul to the 0.2 tube, and 35ul and 25ul to the top and bottom tubes from the 1.2ul (because they were ~350ul, and 250ul respectively)
- Then there was not enough time in the day for another 2 hours in the -80 so I placed them in the -20 overnight
- Then I re-looked at some of the reference protocols and I realized that I should have added 100% ethanol to the tubes after adding the NaOAc!!!! And I realized that adding in that extra NaOAc was probably a bad idea
- I took the tubes out of the freezer and added 2.5 - 2X the volume with 100% ethanol
- ~962ul of 100% ethanol to the 1.2mL top liquid tube
- ~687ul of 100% ethanol to the 1.2mL bottom liquid tube
- 1mL of 100% ethanol to the 0.2mL tube
- Inverted tubes to mix
- The 1.2mL bottom liquid tube immediately became very cloudy
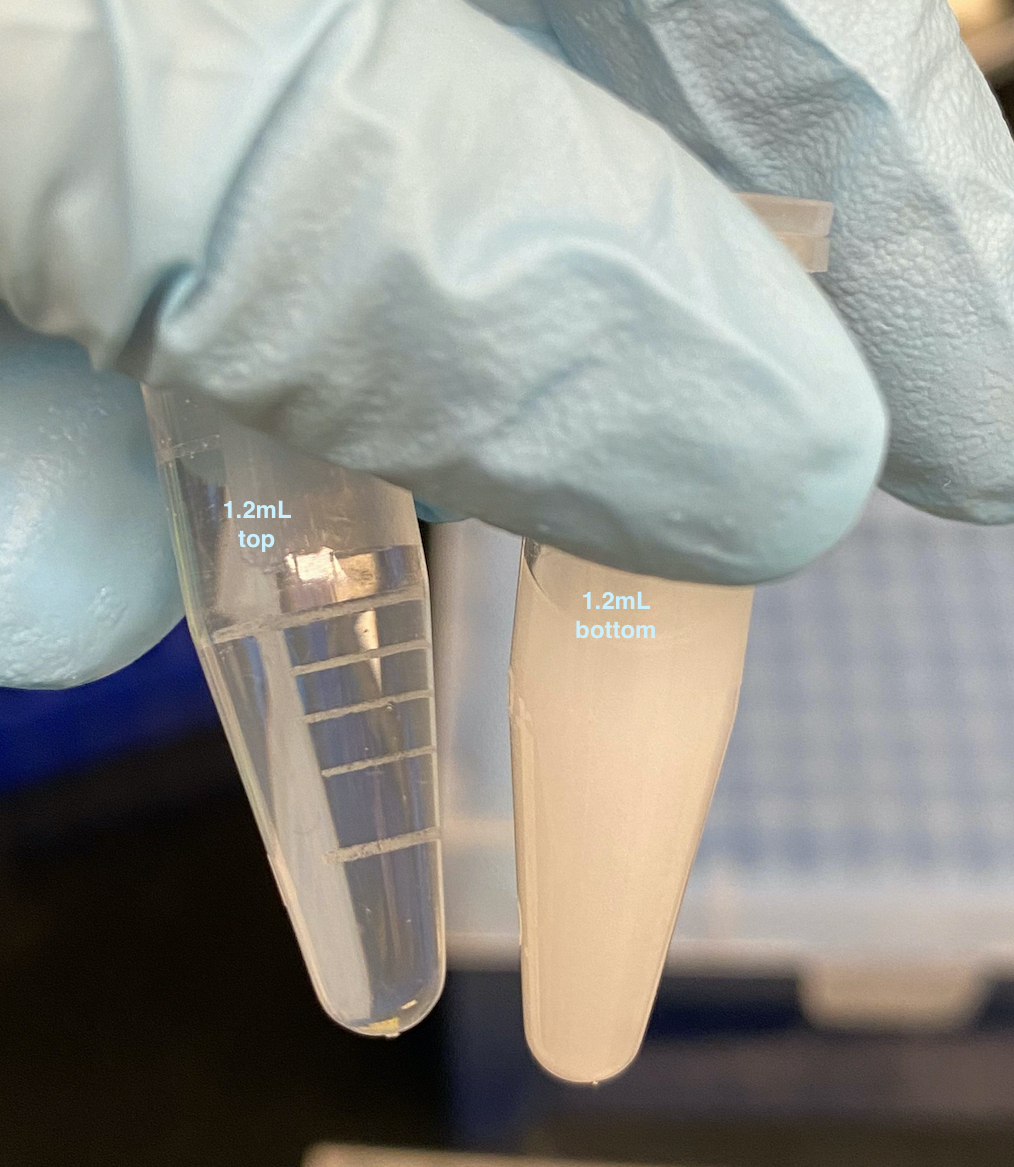
- The 1.2mL bottom liquid tube immediately became very cloudy
- Placed all the tubes back again in the -20 for overnight incubation
20220721 DNA Precipitation Salvage
- Took tubes out of the freezer and centrifuged them in the fridge for 30 minutes at mix speed (16300rcf)
- Decanted off supernatant
- There was a somewhat visible pellet in the 0.2mL tube and 1.2mL top tube
- There was a huge white pellet in the 1.2mL bottom tube
- 0.2mL tube
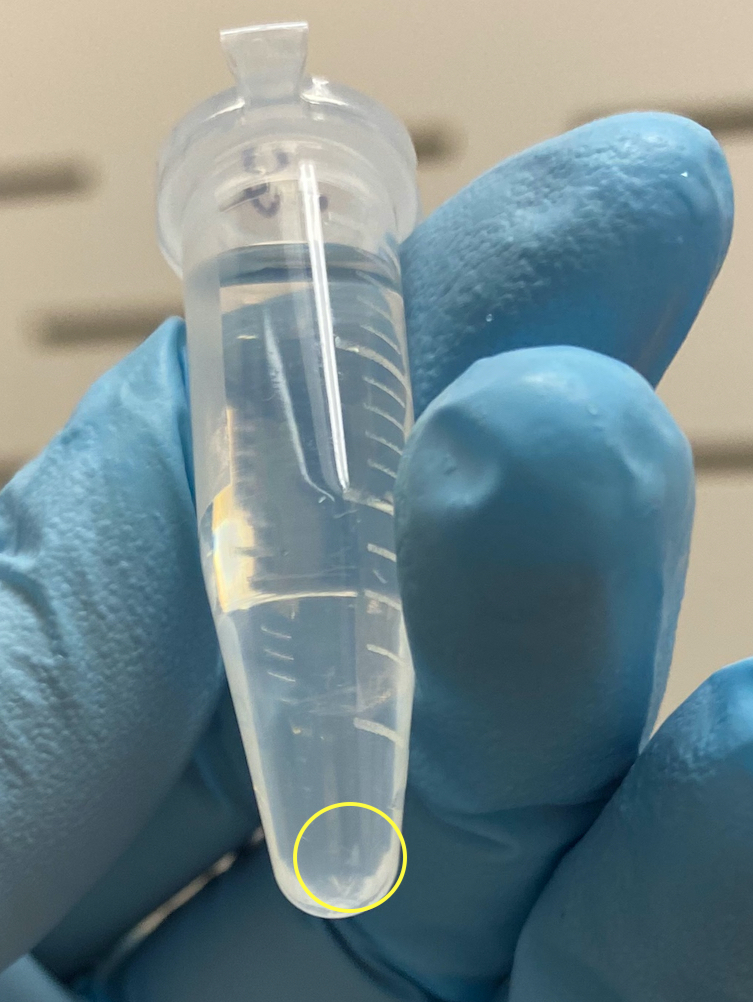
- 1.2mL top tube

- 1.2mL bottom tube

- Added 500ul fresh cold 80% ethanol to each tube
- Centrifuged tubes 30 minutes in the fridge at 16,300rcf
- Removed all ethanol from the tubes (tipped over on kim wipes) and let tubes “dry” a little
- Resuspended pellets in 10mM Tris-HCl
- 1.2mL top: 25ul
- 1.2mL bottom: 200ul
- 0.2mL: 25ul
- Let tubes sit on the bench for a few hours to resuspend
- Qubit BR DNA
- 1.2mL top: 8.73ng/ul
- 1.2mL bottom: 2ng/ul
- 0.2ng/ul: too low to read
Notes
- Obviously I had made a mistake in making my protocol and missed that I needed to add the ethanol after the NaOAc. But it still may have worked for the 1.2mL sample, I will need to do a virus PCR on it and maybe a gel to see the size
- It seems like the more volume/cells is what works better for the extraction
- I want to order phase-lock gel tubes to make removing the the top phase easier after the phenol chloroform