Rethinking the Primers for Regions of DiNV Needed to Amplify Sequences for Cas9 Digestion Tests
I was not able to get either of the 130kb primer pairs to amplify properly after many attempts (see this document). I decided to look into designing new primers for that region, as well as looking into one of the other proposed Cas9 site regions.
Designing new 130kb Primers
- This time, instead of finding primers on my own by trial and error, I used the “design primer” function in Geneious to make primers for me
- I had some specifications that should hopefully make the primers more likely to work this time:
- GC content between 35 and 65
- Tm between 35 and 65
- Max difference in Tms 3 degrees
- Max dimer Tm 30 degrees
- Must have a C or a G at the 3’ end
- No poly-X string longer than 3 bases
- The main things I wanted to add was the C or G at the 3’ end to “hold” the primer down and the restriction on the number of poly-Xs, specifically I had a primer that had a lot of Cs next to each other as one of my original 130kb primers and it may have held on too tight
- I also wanted to make smaller regions to amplify because I wondered if the long PCR product could also be causing me some issues. I was able to get PCR products to amplify for the 77kb region that were ~1.5 and ~2kb, but I wasn’t able to get the 130kb region ~2.5 and ~3kb ones to work
- I want the PCR product to span both Cas9 sites so I can use that product to digest both
- Surprisingly (or not surprisingly because I had a hard time designing primers in the first place), when I selected a close region around the Cas9 sites, Geneious couldn’t find any suitable primers
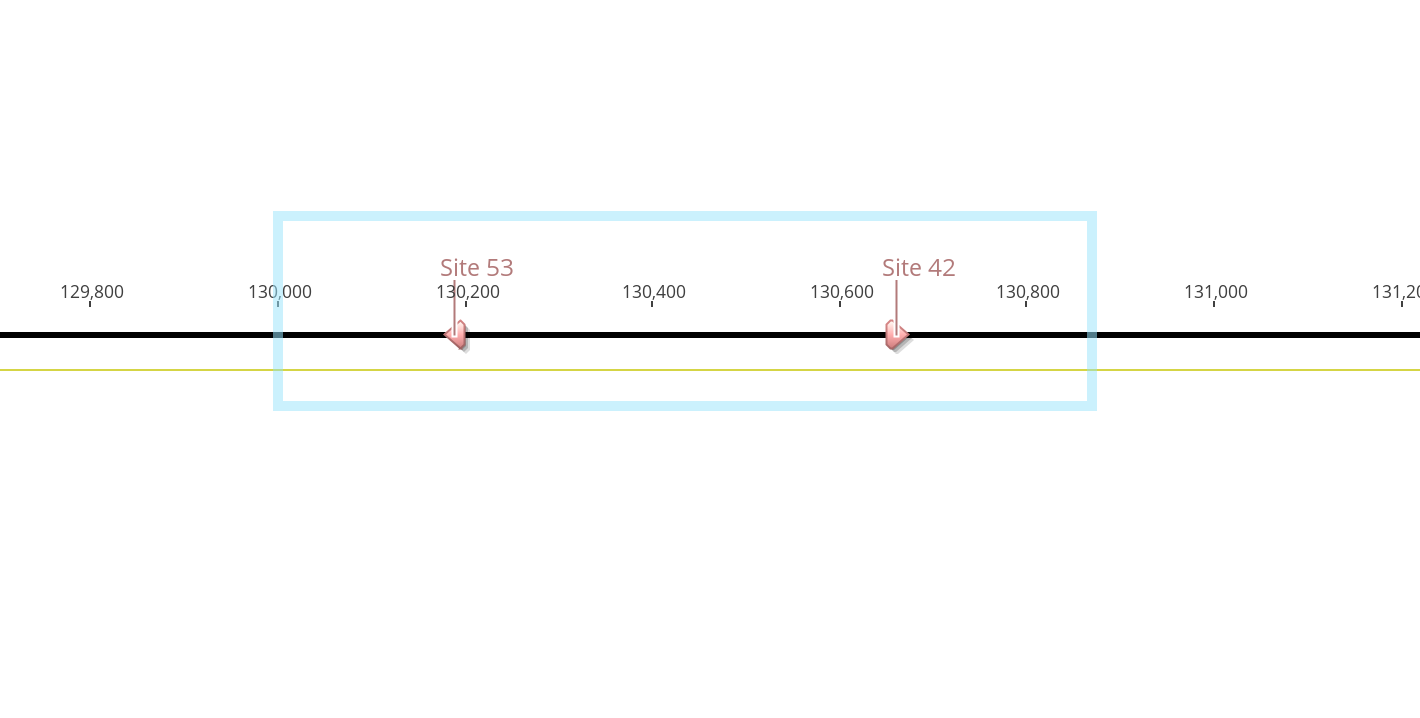
- So I had to expand the region I was looking at. This time it generate 3 primer pairs that fit the parameters

- There are a couple of things about these primers that may not be idea though
- If you can see, primer 131,122 R looks like it’s the same as one I designed previously, 130-DiNV-R-1. There’s actually only a 16bp overlap in them, 131,122 R is a little bit more right than the one I designed. But I worry about something with a very similar primer to one that I already wasn’t able to get to work very well…
- Then there are the 130,860 R and 130,858 R primers. These two have off target sites that the program detected. 130,860 R has an exact match at 46,000 -> 46,019 in DiNV, and 130,860 R (almost exactly the same sequence as 130,860 R) also matches at 46,000 -> 46,019 except for 1 bp that is a mismatch. I think it would be smarter to have the primer with the 1 mismatch
- The off target annealing might not be too bad, because neither 129,666 F, 129,665 F, or 129,778 F have any predicted off target sites
- If we want to go forward with these, the best pairs to get different sized amplicons that aren’t relying on the same primer for both would be 129,778 F - 130,860 R and 129,666 F - 131,122 R. 129,666 F - 131,122 R produced a 1,457bp product. 129,778 F - 130,860 R produces a 1,083bp product
- Sequences:
- 129,778 F GACAAGTTCTGCCACCAACC
- 130,860 R TACCGACAAAGGGATCTAGC (bold letter is the one that mismatches with the 46kb sequence)
- 129,666 F TTCCTACCACGAGCACTACTG
- 131,122 R TGTGGTTTGTGGAGGATAGGC
I began wondering that if I’m having so much trouble designing primers/amplifying the DNA for this region, maybe it is not a good region to try to do the cloning. So I decided to look into the 45kb region again and design primers there. The choice to try the 77kb and 130kb regions was mostly arbitrary.
Designing Primers for 45kb Region
- I went back and looked at the Cas9 sites already investigated for off target sites in D. virilis in the 45kb region and I thought that 45kb-24 and 45kb-44 looked like they might be fine. Neither of them have any BLAST hits to D. virilis with an E. value below 4.8 that include any PAM sequence. Their activity scores are 0.638 and 0.667 respectively
- First I decided to look at the GC content in that region to see if it looked like there was a huge spike in AT% which might represent an origin of replication

- While there is a noticeable dip in GC% right around site 44, it’s very high for site 24, and the dip is similar to surrounding low regions. Additionally, we are assuming that DiNV has multiple origins of replication (genome is so large it would be very unlikely that it would have only 1) that messing up one might not cause us too much issue
- I used the same parameters as above to make the program search for primers
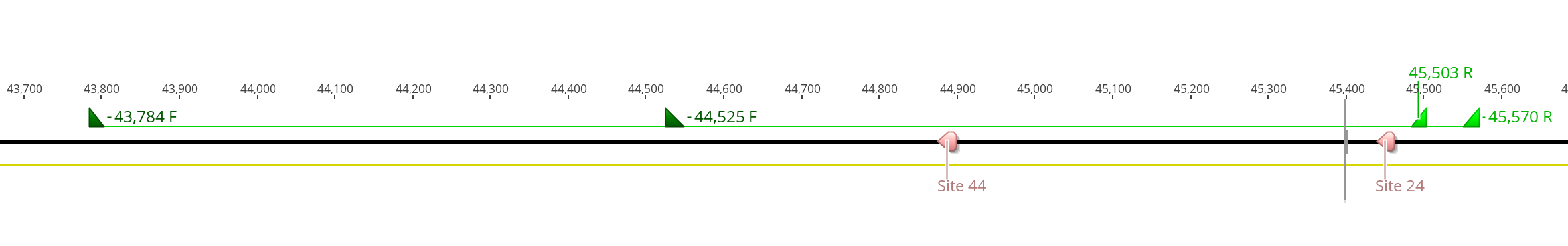
- Again it was kind of hard to get “small” regions to be amplified, the program wouldn’t find any primers that spanned the two Cas9 sites that made a product under 1kb long. But, for these primer pairs there are no expected off target annealings
- 43,784 F - 45,503 R produces a product that is 1,720bp long
- 44,525 F - 45,570 R produces a product that is 1,046bp long
- 43,784 F TGGTGGTGGTCGTTGTGATG
- 45,503 R ACACACCATTCGCTTGTACC
- 44,525 F TCCAACCATGTATGCAATATGACC
- 45,570 R TACTCACACACACCACCACC
PCR Product Digestion Sizes
What would be the expected sizes if these PCR products were digested by the Cas9 guide RNAs?
130kb Region 129,778 F - 130,860 R
- sgRNA 42 cuts at about 130,667
- 130,860 - 130,667 = 198bp
- 130,667 - 129,778 = 889bp
- sgRNA 53 cuts at about 130,182
- 130,860 - 130,182 = 678bp
- 130,182 - 129,778 = 404bp
130kb Region 129,666 F - 131,122 R
- sgRNA 42 cuts at about 130,667
- 131,122 - 130,667 = 455bp
- 130,667 - 129,666 = 1,001bp
- sgRNA 53 cuts at about 130,182
- 131,122 - 130,182 = 940bp
- 130,182 - 129,666 = 516bp
45kb Region 43,784 F - 45,503 R
- sgRNA 24 cuts at about 45,445
- 45,503 - 45,445 = 48bp
- 45,445 - 43,784 = 1,661bp
- sgRNA 44 cuts at about 44,881
- 45,503 - 44,881 = 622bp
- 44,881 - 43,784 = 1,097bp
45kb Region 44,525 F - 45,570 R
- sgRNA 24 cuts at about 45,445
- 45,570 - 45,445 = 125bp
- 45,445 - 44,525 = 920bp
- sgRNA 44 cuts at about 44,881
- 45,570 - 44,881 = 689bp
- 44,881 - 44,525 = 356bp
The only questionable digestion would be for 45kb Region 43,784 F - 45,503 R with sgRNA 24, where it might be hard to see it digest into a 48bp fragment and 1,661bp fragment. Will I be able to see the difference between 1,720bp and 48bp? I might be able to run this one on a second gel with a higher percentage to see the 48bp fragment come out
I also went through an pulled out homology arm sequences from Geneious so we have them ready. All homology arm sequences are here
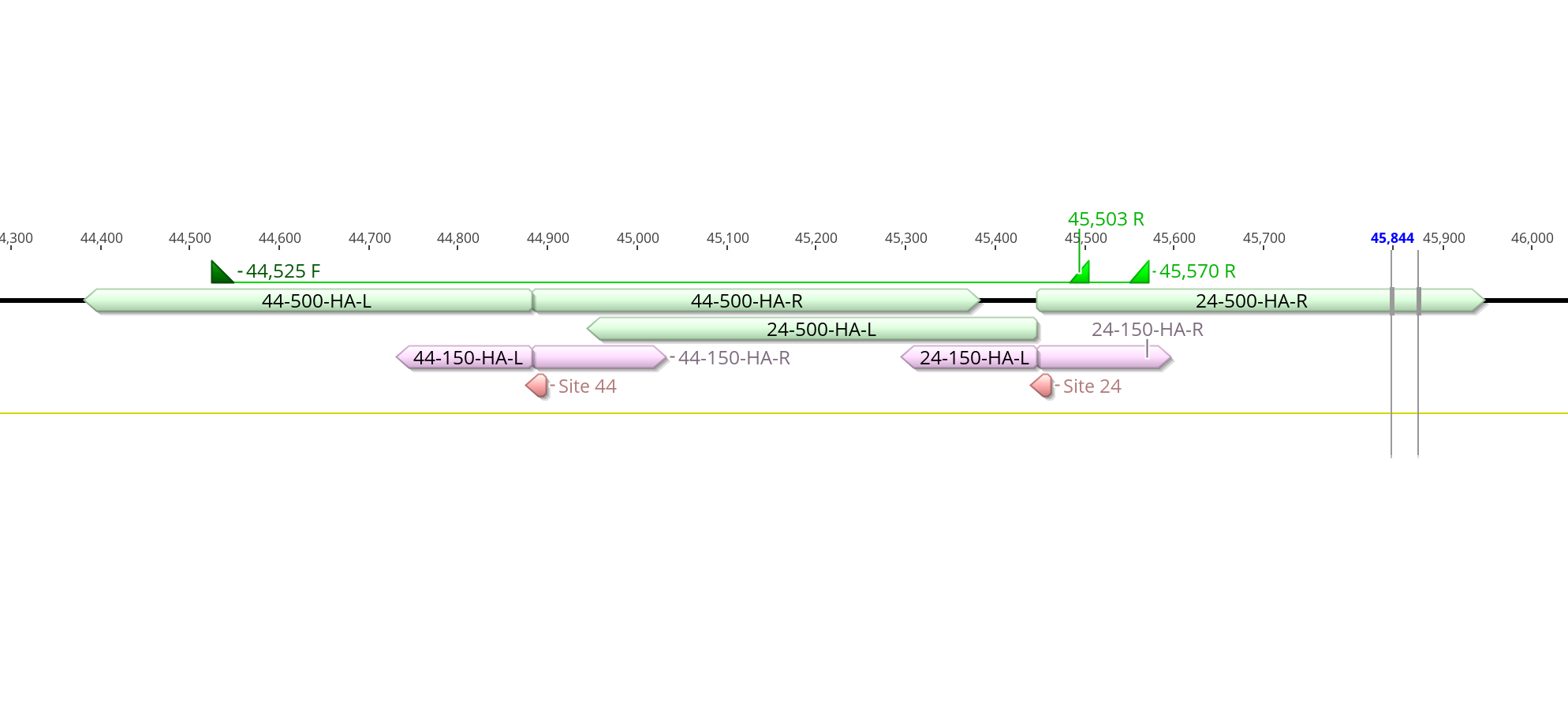
sgRNA primers for the 45kb Cas9 Sites
- Now that I’ve looked into 2 more Cas9 sites, I have to design the primers to make the sgRNAs for them
- Again, I am using the NEB protocol for this
- Select 20 nucleotide target sequence (not including the PAM (NGG) sequence)
- #24 : 5’ GCGCATGTGTATATTTACCA 3’
- #44 : 5’ GGGTGTGTGTCTATGCATTG 3’
- Check input sequence for presence of “G” at the 5´ end. If there are no “G’s” at the 5´ end, add one “G”
- Both have a G on the 5’ end already
- To the 5´ end; append T7 promoter sequence: TTCTAATACGACTCACTATA
- #24 : 5’ TTCTAATACGACTCACTATAGCGCATGTGTATATTTACCA 3’
- #44 : 5’ TTCTAATACGACTCACTATAGGGTGTGTGTCTATGCATTG 3’
- To the 3´ end; append 14 nucleotide overlap sequence: GTTTTAGAGCTAGA
- #24 : 5’ TTCTAATACGACTCACTATAGCGCATGTGTATATTTACCAGTTTTAGAGCTAGA 3’
- #44 : 5’ TTCTAATACGACTCACTATAGGGTGTGTGTCTATGCATTGGTTTTAGAGCTAGA 3’
- Check complete oligo sequence: 5´ TTCTAATACGACTCACTATAG(N)20GTTTTAGAGCTAGA 3´
- #24 : 5’ TTCTAATACGACTCACTATAGCGCATGTGTATATTTACCAGTTTTAGAGCTAGA 3’
- #44 : 5’ TTCTAATACGACTCACTATAGGGTGTGTGTCTATGCATTGGTTTTAGAGCTAGA 3’