Test Amplifying Primers Designed Here on DiNV to be Used for Cas9 Digestion Tests
20220328 Reconsitute Primers to 100uM and Dilute to 10uM for PCR Primers and 1uM for sgRNA Primers
- Primers received from Eurofins Genomics are dried and given a liquid amount to add to the tube for a 100uM stock primer concentration
- Centrifuged all tubes down to be sure dried oligos were at the bottom of the tubes
- Added the appropriate amount of Low TE buffer (low EDTA TE buffer) to each tube to get a 100uM concentration:
- 77-DiNV-F-1: 223ul
- 77-DiNV-R-1: 246ul
- 77-DiNV-F-2: 221ul
- 77-DiNV-R-2: 217ul
- 130-DiNV-F-1: 226ul
- 130-DiNV-R-1: 144ul
- 130-DiNV-F-2: 220ul
- 130-DiNV-R-2: 186ul
- sgRNA-86-77-prim: 225ul
- sgRNA-92-77-prim: 129ul
- sgRNA-42-130-prim: 201ul
- sgRNA-53-130-prim: 208ul
- Vortexed all tubes for 15 seconds, spun down, and kept on ice
- Make 1:10 dilutions for the PCR primers in new tubes
- 90ul molecular grade water
- 10ul 100uM primer
- Vortexed diluted tubes, spun down, and kept on ice
- All tubes stored in -20 freezer in primer box
20220328 PCRs for 77-DiNV-1, 77-DiNV-2, and 130-DiNV-1
77-DiNV-1
- Made a new aliquot of the 2mL HMW DNA sample to be ~10ng/ul
- 38.8ul molecular grade water
- 11.6ul sample
- Decided to try PCRing the newly diluted 2mL sample, the previously diluted 2mL sample, and a negative control (water)
- Made master mix on ice:
- 5mL GoTaq * 3.3 = 16.5ul
- 0.25ul 77-F-1 * 3.3 = 0.825ul
- 0.25ul 77-R-1 * 3.3 = 0.825ul
- 3.5 molec grade water * 3.3 = 11.55ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 54 degrees C 30 sec
- 72 degrees C 1 min 30 sec
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 30 times
- The lowest primer Tm is 59.4 degrees C
- The program ran for 1 hour and 54 minutes
- Tubes were placed in the fridge afterwards
77-DiNV-2
- Used the same samples as above
- Made master mix on ice:
- 5mL GoTaq * 3.3 = 16.5ul
- 0.25ul 77-F-2 * 3.3 = 0.825ul
- 0.25ul 77-R-2 * 3.3 = 0.825ul
- 3.5 molec grade water * 3.3 = 11.55ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 53 degrees C 30 sec
- 72 degrees C 2 minutes
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 30 times
- The lowest primer Tm is 58.9 degrees C
- The program ran for 2 hours and 12 minutes
- Tubes were placed in the fridge afterwards
130-DiNV-1
- Used the same samples as above
- Made master mix on ice:
- 5mL GoTaq * 3.3 = 16.5ul
- 0.25ul 130-F-1 * 3.3 = 0.825ul
- 0.25ul 130-R-1 * 3.3 = 0.825ul
- 3.5 molec grade water * 3.3 = 11.55ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 54 degrees C 30 sec
- 72 degrees C 3 minutes
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 30 times
- The lowest primer Tm is 59.2 degrees C
- The program ran for 2 hours and 12 minutes
- Tubes were placed in the fridge afterwards
20220329 130-DiNV-2 PCR and Gel
130-DiNV-2
- Used the same samples as above
- Made master mix on ice:
- 5mL GoTaq * 3.3 = 16.5ul
- 0.25ul 130-F-2 * 3.3 = 0.825ul
- 0.25ul 130-R-2 * 3.3 = 0.825ul
- 3.5 molec grade water * 3.3 = 11.55ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 54 degrees C 30 sec
- 72 degrees C 2 minutes, 30 seconds
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 30 times
- The lowest primer Tm is 59.2 degrees C
- The program ran for 2 hours and 12 minutes
- Tubes were placed in the fridge afterwards
Ran a 1% gel with all the PCRs at 90V for 40 minutes and stained for 40 minutes. No samples showed up whatsoever.
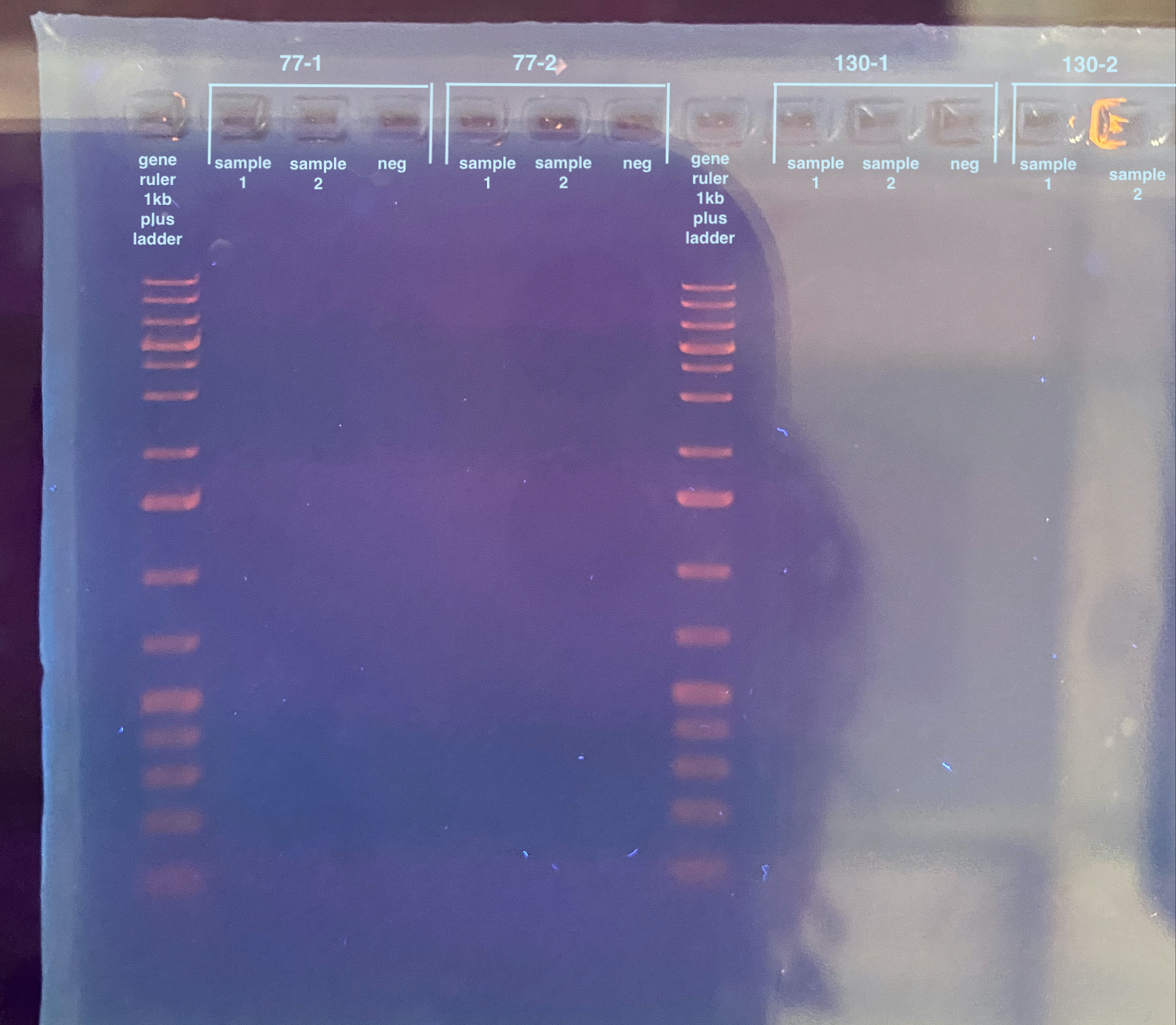
Here I got worried because I designed these primers myself, so if they don’t work it’s truly my fault. But I thought up some ways to try the PCRs again.
20220330 PCR Again of 77-DiNV-1
- Here I changed a few things:
- Lowered the annealing temperature by 2 degrees C to 52 degrees
- Increased the time for annealing from 30 seconds to 1 min
- Increased the cycle number to 34 cycles
- Tested 3 different DNA samples
- Made master mix on ice:
- 5mL GoTaq * 4.4 = 22ul
- 0.25ul 77-F-1 * 4.4 = 1.1ul
- 0.25ul 77-R-1 * 4.4 = 1.1ul
- 3.5 molec grade water * 4.4 = 15.4ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 52 degrees C 1 minute
- 72 degrees C 1 minutes, 30 seconds
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 34 times
- The program ran for 2 hours and 27 minutes
- Tubes were placed in the fridge afterwards until a gel was run
- A 1% gel was run for 40 minutes at 90 volts, then stained for 25 minutes:
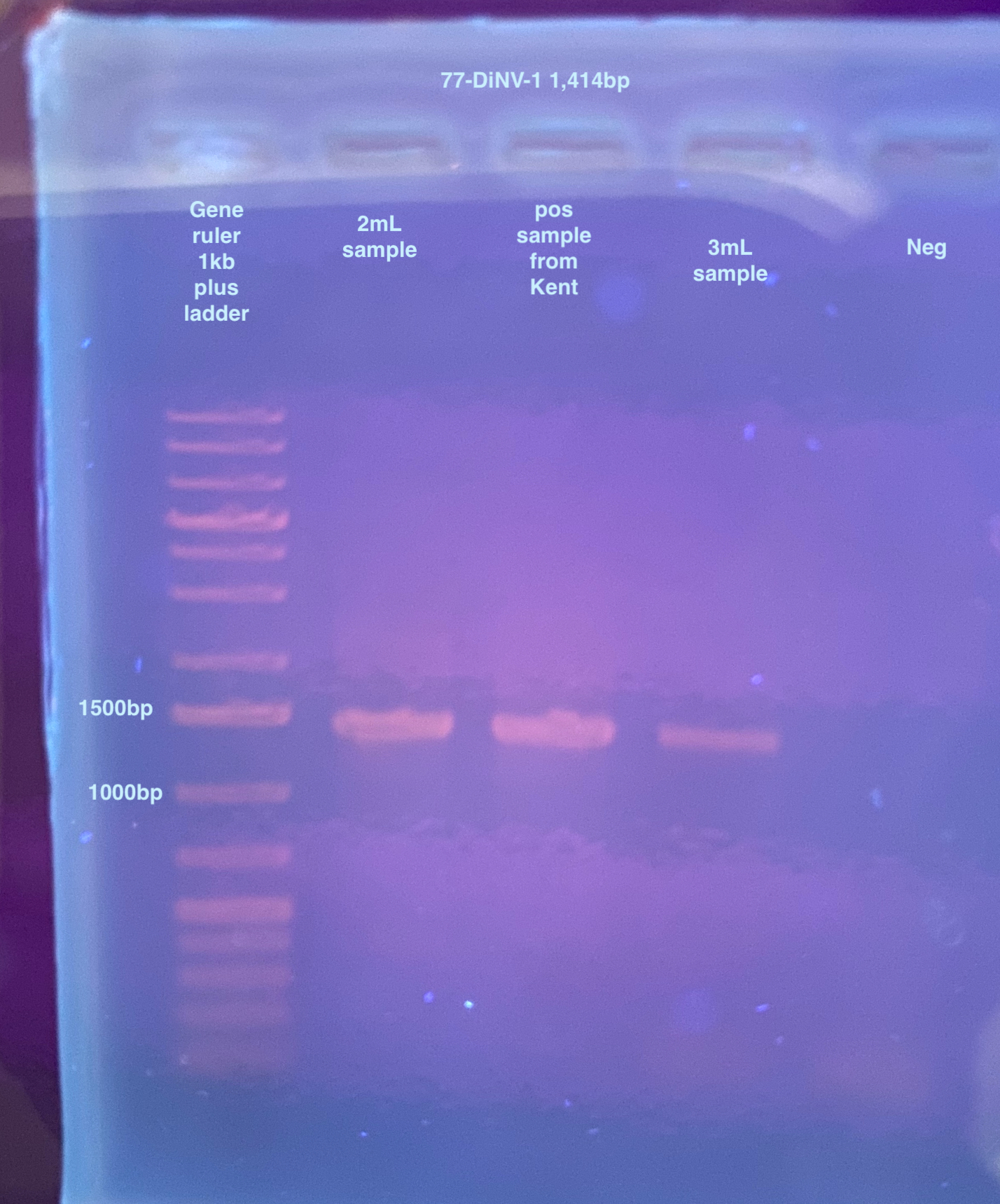
Success! The PCR worked! The product is supposed to be 1,414bp long, and this looks exactly right! Now to try the modified PCR parameters on the other primers
20220331 77-DiNV-2 PCR Again
- Here I only used the 2mL sample as the sample because I know it should work
- Made master mix on ice:
- 5mL GoTaq * 2.2 = 11ul
- 0.25ul 77-F-2 * 2.2 = 0.55ul
- 0.25ul 77-R-2 * 2.2 = 0.55ul
- 3.5 molec grade water * 2.2 = 7.7ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 52 degrees C 1 minute
- 72 degrees C 2 minutes
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 34 times
- The program ran for 2 hours and 44 minutes
- Tubes were placed in the fridge afterwards
20220401 130-DiNV-1 PCR Again
- Here I only used the 2mL sample as the sample because I know it should work
- Made master mix on ice:
- 5mL GoTaq * 2.2 = 11ul
- 0.25ul 130-F-1 * 2.2 = 0.55ul
- 0.25ul 130-R-1 * 2.2 = 0.55ul
- 3.5 molec grade water * 2.2 = 7.7ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 52 degrees C 1 minute
- 72 degrees C 3 minutes
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 34 times
- The program ran for 3 hours and 20 minutes
- Tubes were placed in the fridge afterwards
20220401 130-DiNV-2 PCR Again
- Here I only used the 2mL sample as the sample because I know it should work
- Made master mix on ice:
- 5mL GoTaq * 2.2 = 11ul
- 0.25ul 130-F-2 * 2.2 = 0.55ul
- 0.25ul 130-R-2 * 2.2 = 0.55ul
- 3.5 molec grade water * 2.2 = 7.7ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 52 degrees C 1 minute
- 72 degrees C 2 minutes 30 seconds
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 34 times
- The program ran for 3 hours and 2 minutes
- Tubes were placed in the fridge afterwards
20220404 Gel
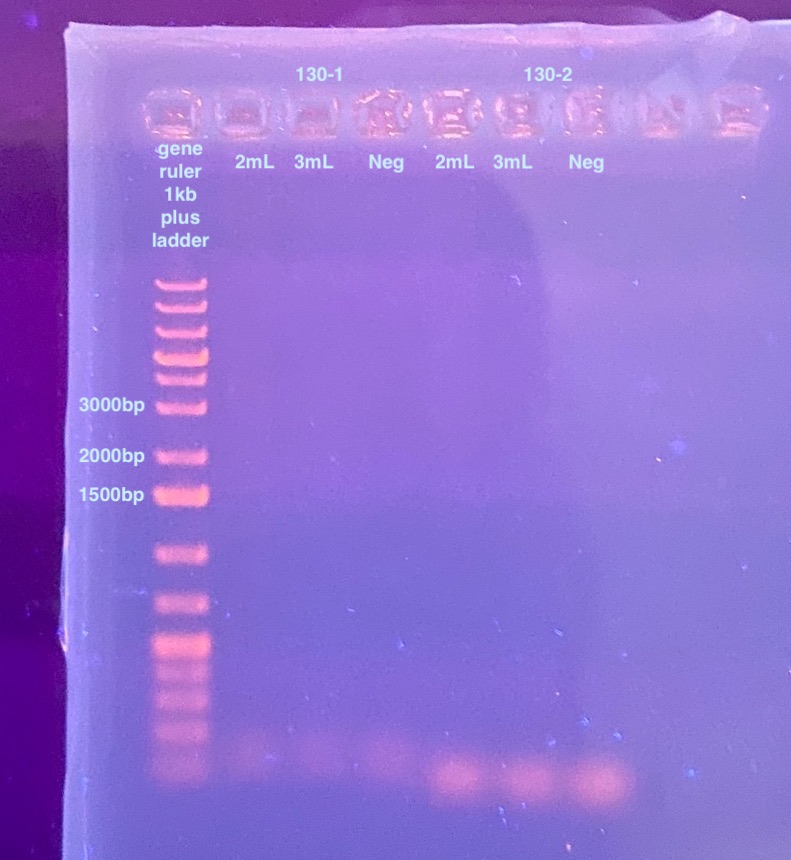
- The 77-2, 130-1, and 130-2 PCRs were run on a 1% gel for 35 minutes at 90V

There is proper amplification of 77-DiNV-2, but none for either of the 130 regions. There is a small amount of off target amplification for the 77 amplicon, I can hopefully get rid of that by lowering the primer concentration
20220405 Retry 130 1 and 2
- I tried both the 2mL sample and the 3mL sample, and lowered the annealing temp to 50 degrees C
- 130-DiNV-1
- Made master mix on ice:
- 5mL GoTaq * 3.3 = 16.5ul
- 0.25ul 130-F-1 * 3.3 = 0.825ul
- 0.25ul 130-R-1 * 3.3 = 0.825ul
- 3.5 molec grade water * 3.3 = 11.55ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 50 degrees C 1 minute
- 72 degrees C 3 minutes
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 34 times
- Tubes were placed in the fridge afterwards
- 130-DiNV-2
- Made master mix on ice:
- 5mL GoTaq * 3.3 = 16.5ul
- 0.25ul 130-F-2 * 3.3 = 0.825ul
- 0.25ul 130-R-2 * 3.3 = 0.825ul
- 3.5 molec grade water * 3.3 = 11.55ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 50 degrees C 1 minute
- 72 degrees C 2 minutes and 30 seconds
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 34 times
- Tubes were placed in the fridge afterwards
- Then a 1% gel was run for 35 minutes at 90V:
Again no proper amplification…
20220405 Retry 130-DiNV-2 with Gradient
- I talked to Kistie and she helped me set up a gradient PCR on the machine to try different annealing temperatures
- She also said that, at least for the 130-DiNV-2 primer, it may have too many Cs and hold on too tight to the DNA, and that I should lower my annealing time to 30 seconds
- So I decided to do a gradient from 40 degrees to 50 degrees and put samples at a couple of different annealing temperatures:
- 40 degrees - tubes A
- 41.9 degrees - tubes B
- 43.9 degrees - tubes C
- 46.2 degrees - tubes D
- 47.9 degrees - tubes E
- 49.2 degrees - tubes F
- Each of these temperatures will get a 2mL sample and a negative control
- Made master mix on ice:
- 5mL GoTaq * 13 = 65ul
- 0.25ul 130-F-2 * 13 = 3.25ul
- 0.25ul 130-R-2 * 13 = 3.25ul
- 3.5 molec grade water * 13 = 45.5ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- gradient degrees C 30 seconds
- 72 degrees C 2 minutes and 30 seconds
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 34 times
- Tubes were placed in the fridge afterwards
- Then a 1% gel was run for 35 minutes at 90V:

Basically nothing amplified in any of these, except I saw something in the 40 degree sample. It was very faint and didn’t show up well in the picture. I want to try more cycles with that temp
20220407 Retry 130-DiNV-2 with 40 cycles
- Made master mix on ice:
- 5mL GoTaq * 2.2 = 11ul
- 0.25ul 130-F-2 * 2.2 = 0.55ul
- 0.25ul 130-R-2 * 2.2 = 0.55ul
- 3.5 molec grade water * 2.2 = 7.7ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tube
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- 40 degrees 30 seconds
- 72 degrees C 2 minutes and 30 seconds
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 40 times
- Tubes were placed in the fridge afterwards
- Then a 1% gel was run for 35 minutes at 90V:
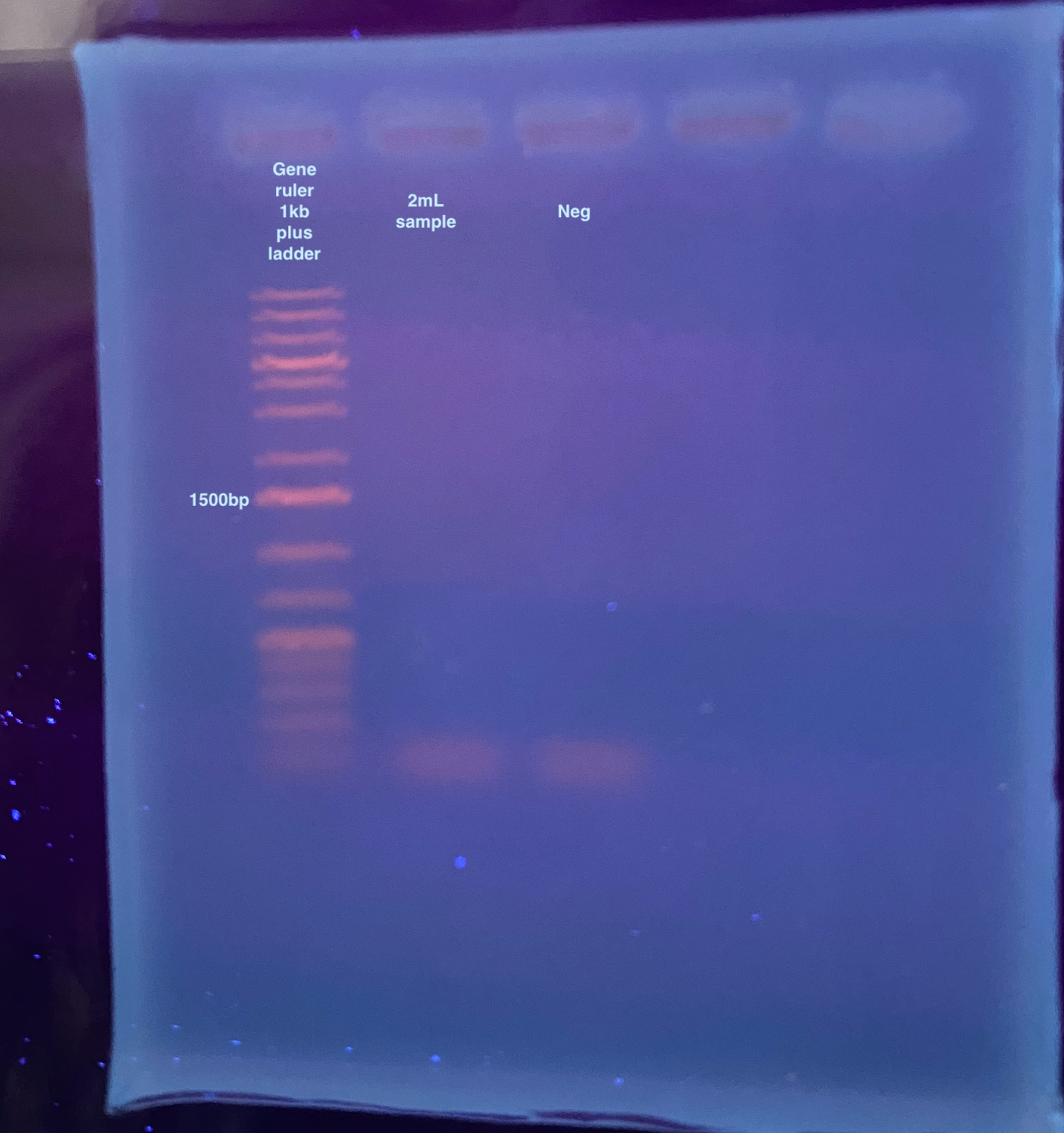
There is nothing that showed up, honestly I probably got a tiny faint thing show up in the 20220405 gel because it stained for 1.5hours.
At this point I stopped trying to get 130-DiNV-2 to work because it wouldn’t amplify in any circumstance. I decided to try the same gradient on 130-DiNV-1.
20220411 Try 130-DiNV-1 with Gradient
- Trying the same gradient as above:
- 40 degrees - tubes A
- 41.9 degrees - tubes B
- 43.9 degrees - tubes C
- 46.2 degrees - tubes D
- 47.9 degrees - tubes E
- 49.2 degrees - tubes F
- Each of these temperatures will get a 2mL sample and a negative control
- Made master mix on ice:
- 5mL GoTaq * 13 = 65ul
- 0.25ul 130-F-2 * 13 = 3.25ul
- 0.25ul 130-R-2 * 13 = 3.25ul
- 3.5 molec grade water * 13 = 45.5ul
- Vortexed and spun down mix, kept on ice
- Assembled PCR strip tubes on ice:
- Added 9ul master mix to each tube
- Added 1ul DNA to the sample tubes
- Added 1ul molec grade water to the negative control tube
- Vortexed and spun down tubes
- Placed tubed in the thermocycler:
- 95 degrees C 2 min
- 95 degrees C 30 sec
- gradient degrees C 30 seconds
- 72 degrees C 3 minutes
- 72 degrees C 5 min
- 12 degrees C hold
- Italic lines are cycled 34 times
- Tubes were placed in the fridge afterwards
- Then a 1% gel was run for 35 minutes at 90V:
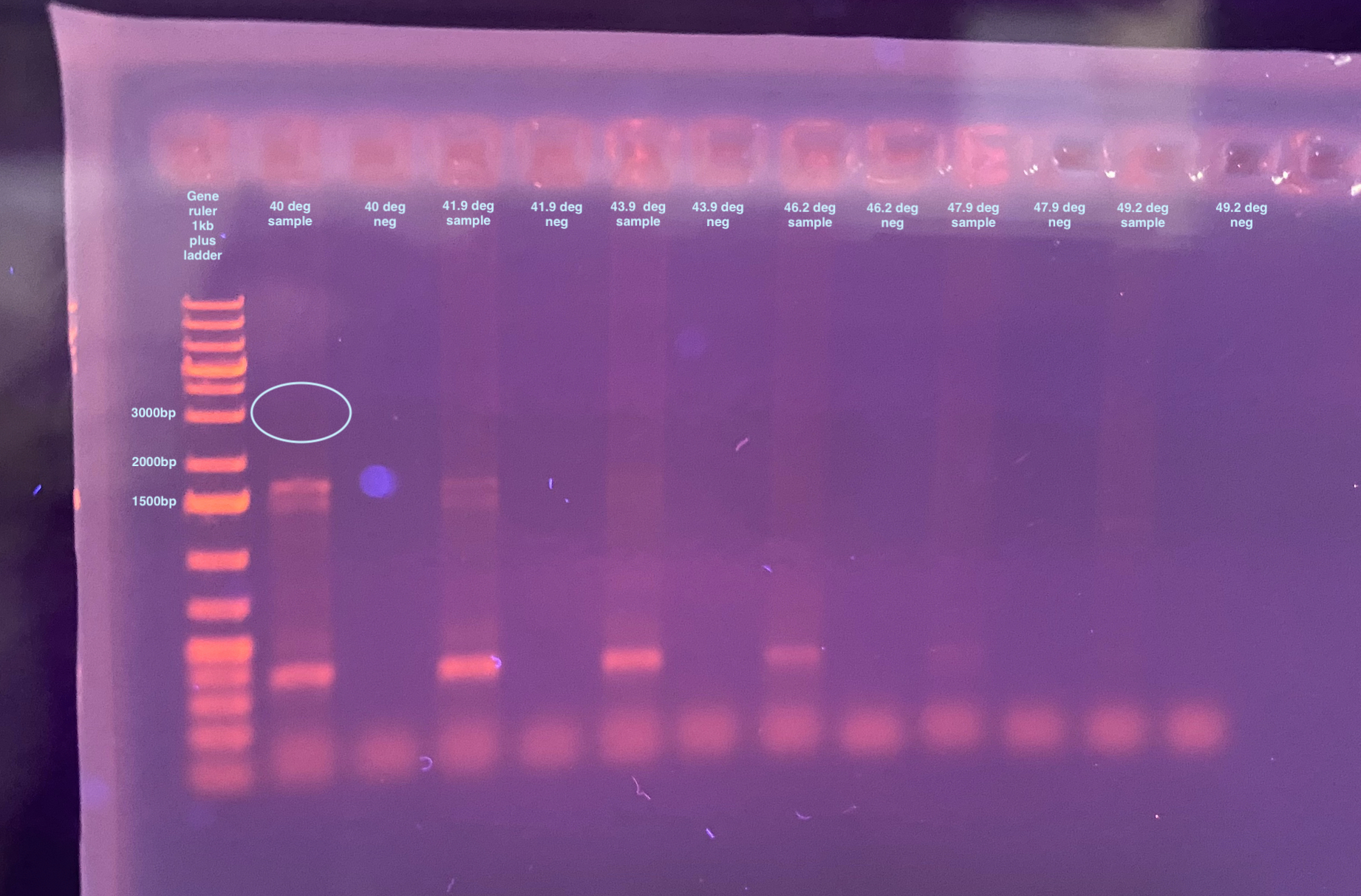
Well here I got some things to amplify but they are all off target. The product is supposed to be ~3,000bp long. I might have to try making primers for this region again and making them amplify a smaller region.