Re-Checking Guide RNA Sites in DiNV Genome
sgRNAs and their BLAST results to the DiNV genome
BLAST parameters:
- Using the sgRNA sequence with the PAM site
- Using D. virilis genome
- Using RegSeq genome database only
- Filter to e value of 0 to 10 only (or if there were more than 20, only looked at between 0 and 1)
Then: Look through each entry and see if there are any BLAST hits that include the 23bp, meaning that the entire PAM site is there, which means there is a chance of the Cas9 cutting.
| Site set # | bp region | Site # | activity score | direction | bp with PAM | BLAST GGG | BLAST TGG | BLAST AGG | BLAST CGG | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 77kb | 86 | 0.713 | reverse | 5’ ACACAAGTGTTGTTATACGATGG 3’ | No hits with below 10 E value including GGG PAM | 1 hit with 4.8 E value, 14/14 identity, includes TGG PAM | No hits with below 10 E value including AGG PAM | No hits with below 10 E value including CGG PAM | |
| 1 | 77kb | 92 | 0.719 | forward | 5’ AAAAAATGCACTAAAACACAGGG 3’ | No hits with below 10 E value including GGG PAM | No hits with below 10 E value including TGG PAM | 1 hit with 4.8 E value, 17/18 identity, AGG PAM | No hits with below 10 E value including CGG PAM | |
| 1 | 77kb | 27 | 0.716 | reverse | 5’ TGTATTGTCAGTGTGGGAGATGG 3’ | 1 hit with 0.077 E value, 17/17 identity, GGG PAM. 3 hits with 4.8 E value, 14/14 identity, GGG PAM. | 1 hit with 0.077 E value, 17/17 identity, includes TGG PAM. 2 hits with 4.8 E value, 14/14 identity, includes PAM | 3 hits with 4.8 E value, 14/14 identity, includes PAM | 1 hit with 4.8 E value, 14/14 identity, includes PAM | |
| 1 | 77kb | 73 | 0.623 | reverse | 5’ ATCTAAATAAATACAATCGAGGG 3’ | No hits with below 10 E value including the GGG PAM | No hits with below 10 E value including TGG PAM | No hits with below 10 E value including AGG PAM | 1 hit with 1.2 E value, 15/15 identity, includes CGG PAM | |
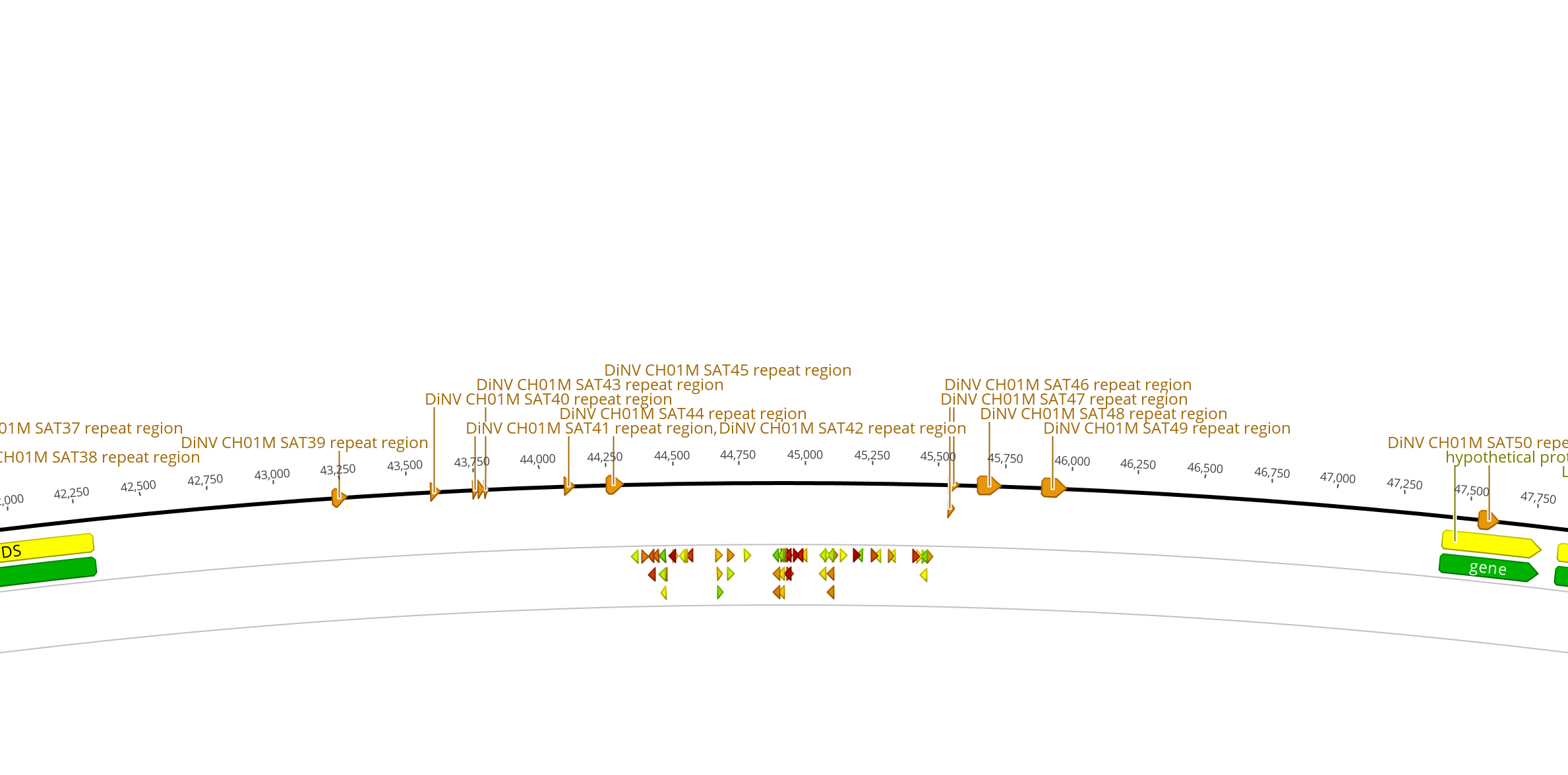
| 2 | 45kb | 44 | 0.667 | reverse | 5’ GGGTGTGTGTCTATGCATTGGGG 3’ | 1 hit at 4.8 E. value, 20/22bp identity, includes GGG PAM. | A few with 4.8 E value, 14/14 identity, includes CGG PAM (See image) | No hits with below 10 E value including AGG PAM | No hits with below 10 E value including CGG PAM | |
| 2 | 45kb | 54 | 0.662 | reverse | 5’ TGTGTGTGTGTGTAGTTGGGGGG 3’ | 2 hits with 0.005 E value 22/23bp identity, includes GGG PAM. | very repetitive, might not be good | |||
| 2 | 45kb | 24 | 0.638 | reverse | 5’ GCGCATGTGTATATTTACCAGGG 3’ | 2 hits at 4.8 E value, 14/14 identity, GGG PAM | No hits with below 10 E value including TGG PAM | 1 hit with 4.8 E vlaue, 14/14 identity, includes PAM | No hits with below 10 E value including CGG PAM | |
| 2 | 45kb | 27 | 0.632 | reverse | 5’ CGCAACACACCAAAATACGATGG 3’ | 1 hit with 4.8 E vlaue, 14/14 identity, includes PAM | 1 hit at 1.2 E value, 15/15 identity, includes TGG PAM, 1 hit at 4.8 E value, 14/14 identity, includes TGG PAM | No hits with below 10 E value including AGG PAM | No hits with below 10 E value including CGG PAM | |
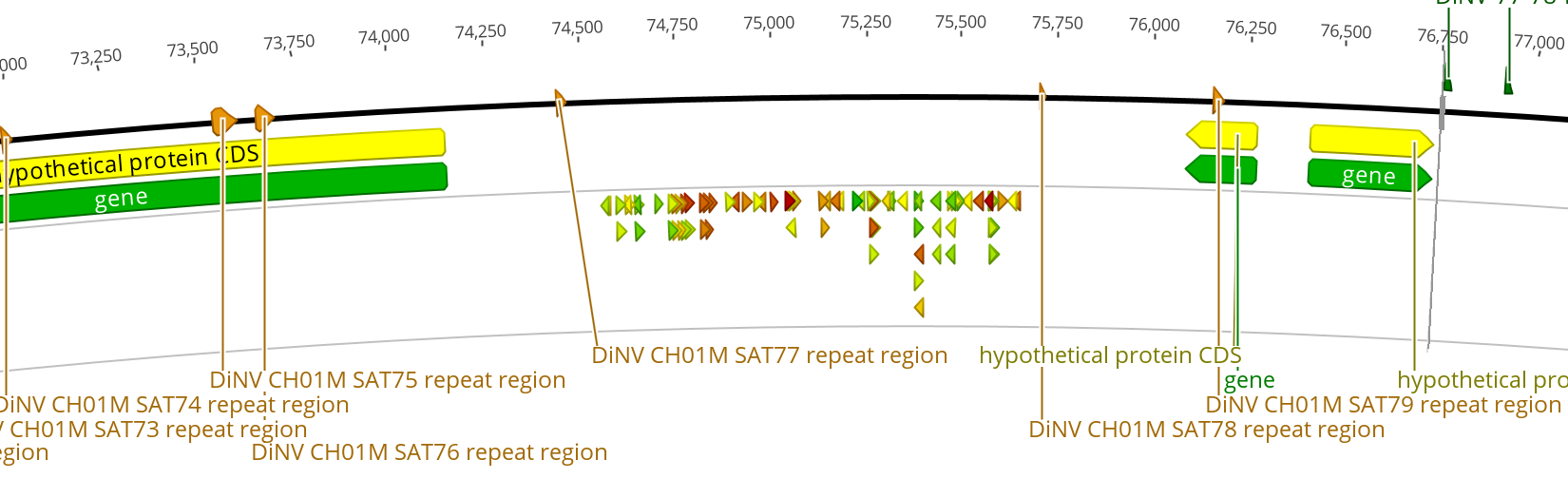
| 3 | 75kb | 32 | 0.784 | forward | 5’ GTGACCATACACACACACAGTGG 3’ | None with E value below 1 (too many others to go through) | 1 hit at 0.005 E value, 19/19 identity, includes TGG PAM. 1 hit at 0.02 E value, 18/18 identity, TGG PAM | No hits with below 10 E value including the AGG PAM | No hits with below 10 E value including the CGG PAM | |
| 3 | 75kb | 42 | 0.718 | forward | 5’ CAACTCGATAGAAGTCGACGGGG 3’ | 1 hit at 4.8 E value, 14/14 identity, includes GGG PAM. | 1 hit with 0.077 E value, 17/17 identity, includes TGG PAM | 3 hits at 4.8 E value, 14/14 identity, includes AGG PAM | No hits with below 10 E value including the CGG PAM | very few results |
| 3 | 75kb | 61 | 0.683 | reverse | 5’ TGTTCGAGAGAGAGATTGAGGGG 3’ | 1 hit at 0.077 E value, 17/17 identity, including GGG PAM. 2 hits at 0.3 E value, 16/16 identity, including GGG PAM | 1 hit at 1.2 E value, 18/19 identity, includes TGG PAM. 5 hits at 4.8 E value, 17/18 identity, includes TGG PAM. | 1 hit at 1.2 E value, 15/15 identity, includes AGG PAM. 2 hits at 4.8 E value, 17/18 identity, includes AGG PAM. 1 hit at 4.8 E value, 14/14 identity, include AGG PAM | 2 hits at 4.8 E value, 17/18 identity, include CGG PAM | |
| 3 | 75kb | 40 | 0.682 | forward | 5’ CCCAACTCGATAGAAGTCGACGG 3’ | No hits with below 10 E value including GGG PAM | No hits with below 10 E value including TGG PAM | 1 hit at 4.8 E value, 14/14 identity, AGG PAM | 1 hit at 4.8 E value, 14/14 identity, CGG PAM | Very few results |
| 3 | 75kb | 78 | 0.679 | reverse | 5’ ACTAGTCTAACACTATTCCGAGG 3’ | No hits with below 10 E value including GGG PAM | No hits with below 10 E value including TGG PAM | No hits with below 10 E value including AGG PAM | No hits with below 10 E value including CGG PAM | No PAM hits at all! |
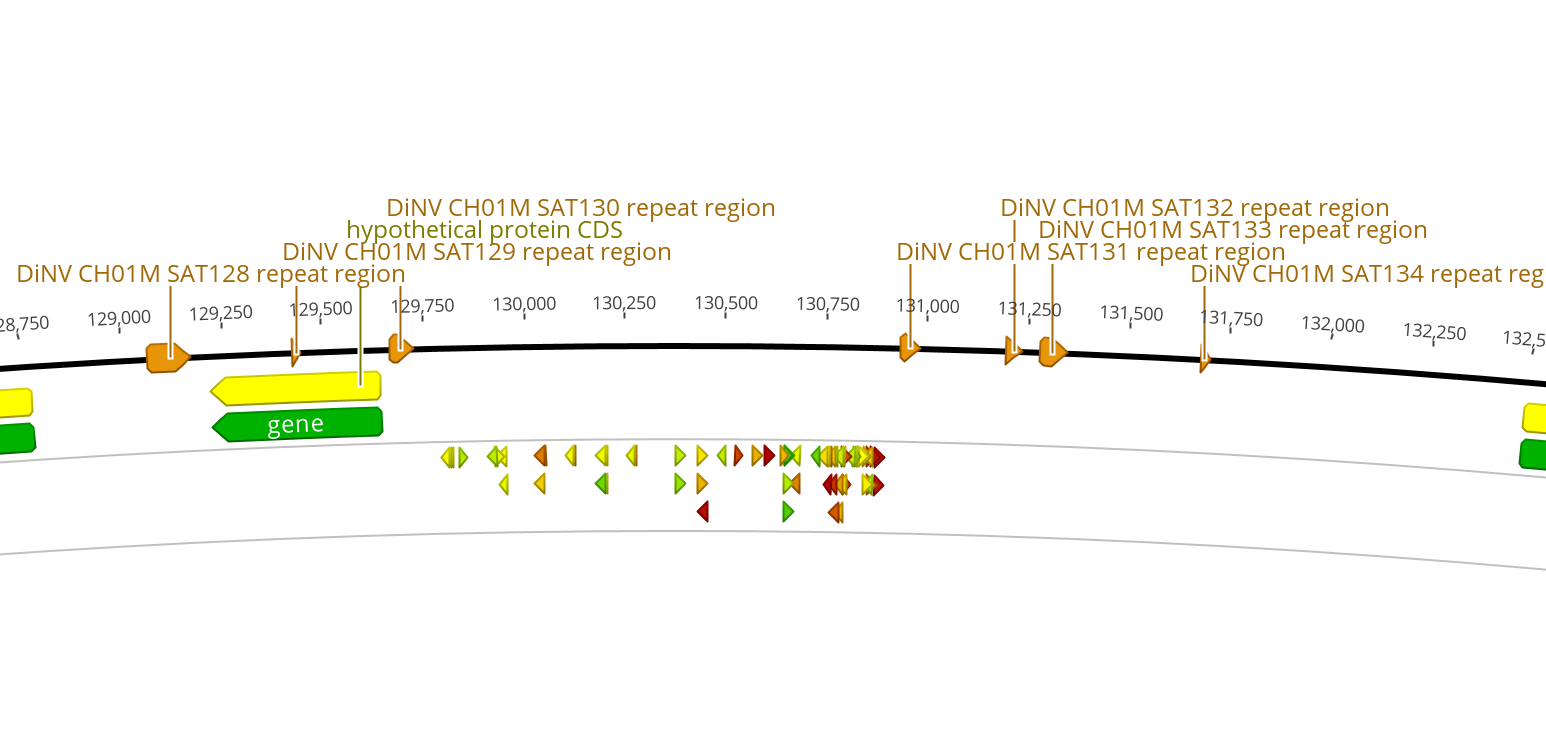
| 4 | 130kb | 40 | 0.713 | forward | 5’ CGTCGATATTGGTCACCCAGAGG 3’ | No hits with below 10 E value including GGG PAM | No hits with below 10 E value including TGG PAM | No hits with below 10 E value including AGG PAM | 1 hit at 4.8 E value, 14/14 identity, CGG PAM | very few results |
| 4 | 130kb | 42 | 0.701 | forward | 5’ TCGATATTGGTCACCCAGAGGGG 3’ | 1 hit at 4.8 E value, 14/14 identity, GGG PAM | No hits with below 10 E value including TGG PAM | 2 hits at 1.2 E value, 15/15 identity, includes AGG PAM | No hits with below 10 E value including CGG PAM | |
| 4 | 130kb | 22 | 0.675 | reverse | 5’ AAAATGGCAAAAATCGAGCTCGG 3’ | 1 hit at 4.8 E value, 14/14 identity, GGG PAM | No hits with below 10 E value including TGG PAM | No hits with below 10 E value including AGG PAM | 1 hit at 4.8 E value, 14/14 identity, CGG PAM. | |
| 4 | 130kb | 43 | 0.667 | forward | 5’ CGATATTGGTCACCCAGAGGGGG 3’ | 1 hit at 4.8 E value, 14/14 identity, GGG PAM | No hits with below 10 E value including TGG PAM | No hits with below 10 E value including AGG PAM | No hits with below 10 E value including CGG PAM | very few results |
| 4 | 130kb | 53 | 0.666 | reverse | 5’ ATACATATACACTTGATGGGTGG 3’ | No hits with below 10 E value including GGG PAM | 1 hit at 4.8 E value, 14/14 identity, TGG PAM | No hits with below 10 E value including AGG PAM | No hits with below 10 E value including CGG PAM |
Characteristics of sgRNA locations
77kb Region (original from grant): site set 1
- Intergenic region: 1,727bp
- No repeats within region
- GC% in intergenic region: 26%
- Flanking genes:
- gp053 189bp
- gp054 237bp
- gp094-like 330bp

45kb Region: site set 2
- Intergenic region used to find CRISPR sites: 1,198bp
- Entire intergenic region between closest genes: 5,081bp
- 2,018bp on left side
- 1,904bp on the right side
- GC% in entire intergenic region: 28%
- 10 repeats regions within the larger region, but none within the CRISPR site region
- Flanking genes:
- gp30 900bp
- gp31 378bp
- gp32 2,940bp
75kb Region: site set 3
- Intergenic region used to find CRISPR sites: 1,234bp
- Entire intergeic region between closest genes: 1,948bp
- 389bp left side
- 424bp right side
- GC% in entire intergenic region: 32%
- 2 repeat regions within the larger region, but none within the CRISPR site region
- Flanking genes:
- gp52 1,22bp
- gp053 189bp
- gp054 237bp
130kb Region: site set 4
- Intergenic region used to find CRISPR sites: 1,189bp
- Entire intergenic region between closest genes: 2,822bp
- 123bp left side
- 1589bp right side
- GC% in entire intergenic region: 30%
- 5 repeat regions within the larger region, but none within the CRISPR site region
- Flanking genes:
- gp88 657bp
- gp89 426bp
- gp90 2748bp