PCR RPL11 (Nuclear Genome Control) on Gel Extraction Products from Zymoclean Gel Extraction and HMW Extracts, As Well As p47 PCR on HMW Extracts to Check for Presence of Virus in HMW Extracts
Notes
- Using qPCR primers from Kent for RPL11 (a ribosomal component)
- Not sure what the PCR conditions should be, not used before on our machines, and I wasn’t able to find anything other than qPCR cycling conditions in papers I looked up. I decided to make the program based off of the Tm and the expected size. The lowest Tm is 62.4, so I used an annealing temp of 57. I guessed that the product would be the full gene (this turned out to be incorrect, it’s a very small part of the gene), so I looked it up and RPL11 in D. virilis is ~1.4kb long. So I did a long extention time in the PCR. This may be unecessary, but it worked so I no longer want to change the PCR program
RPL11 20211206
- Running the 4 “3mL” samples from the gel extraction, plus the 3mL and 2mL HMW extractions, and a pos and neg control: 8 samples, n = 8.8
- Needed to create a diluted stock of primers for myself:
- 10ul vir_RPL11_F primer and 90ul molec grade water
- 10ul vir_RPL11_R primer and 90ul molec grade water
- Needed to dilute the HMW extraction samples because they are way too concentrated to use in a PCR
- 3mL-120hr is 115ng/ul: dilute 2ul in 28ul TE buffer
- 2mL-120hr is 43ng/ul: dilute 1.2ul in 18.8ul
- Using 4ul DNA per sample, except for the positive control, ngeative control, and the 3mL-120hr sample
- Created master mix on ice:
- 1ul 10X NEB taq buffer * 8.8 = 8.8ul
- 1ul 2mM dNTPs * 8.8 = 8.8ul
- 1ul 25mM MgCl2 * 8.8 =8.8ul
- 0.25ul 10uM vir_RPL11_F primer * 8.8 = 2.2ul
- 0.25ul 10uM vir_RPL11_R primer * 8.8 = 2.2ul
- 0.1ul NEB Taq * 8.8ul = 0.88ul
- 2.4ul molecular grade water * 8.8 = 21.12ul
| tube # | sample | ul master mix | ul DNA | ul molec grade water |
|---|---|---|---|---|
| 1 | 3mL-150 | 6 | 4 | 0 |
| 2 | 3mL-UPPER | 6 | 4 | 0 |
| 3 | 3mL-LOWER | 6 | 4 | 0 |
| 4 | 3mL-WELL | 6 | 4 | 0 |
| 5 | 3mL-120hr | 6 | 2 | 2 |
| 6 | 2mL-120hr | 6 | 4 | 0 |
| 7 | pos control | 6 | 1 | 3 |
| 8 | neg control | 6 | 0 | 4 |
- Vortexed and spun down and placed in PCR program RPL11:
- 95 degrees C 3 minutes
- 95 degrees C 30 seconds
- 57 degrees C 1 minute
- 68 degrees C 1 minute 30 seconds
- 68 degrees C 5 minutes
- 12 degree C hold
- bold sections are cycled 34 times
- After, tubes were put in the fridge overnight until the gel
p47 20211206
- Using only the 3mL-120hr, 2mL-120hr, and pos and neg control samples for this PCR, just to test if p47 amplifies in my original HMW extractions
- Made master mix on ice:
- 1ul 10X NEB buffer * 4.4 = 4.4ul
- 1ul 2mM dNTPs * 4.4 = 4.4ul
- 1ul 25mM MgCl2 * 4.4 = 4.4ul
- 0.25ul p47_F * 4.4 = 1.1ul
- 0.25ul p47_R * 4.4 = 1.1ul
- 0.1ul NEB taq * 4.4 = 0.44ul
- 2.4ul molecular grade water * 4.4 = 10.56ul
- Using the same DNA addition method as with RPL11 above
| tube # | sample | ul master mix | ul DNA | ul molec grade water |
|---|---|---|---|---|
| 1 | 3mL-120hr | 6 | 2 | 2 |
| 2 | 2mL-120hr | 6 | 4 | 0 |
| 3 | neg control | 6 | 0 | 4 |
| 4 | pos control | 6 | 1 | 3 |
- Vortexed and spun down and put in the PCR program for p47
- 94 degrees C 5 minutes
- 94 degrees C 45 seconds
- 55 degrees C 1 minute
- 68 degrees C 1.5 minutes
- 68 degrees C 5 minutes
- Hold at 12 degrees C
- bold text is cycled through 30 times
- After, tubes were put in the fridge for overnight until a gel could be run
1% gel 20211207
- 1% gel: 1g agarose and 100mL 1X TAE
- Ran for 40 minutes at 90V
- Incubated in EtBr for ~60min
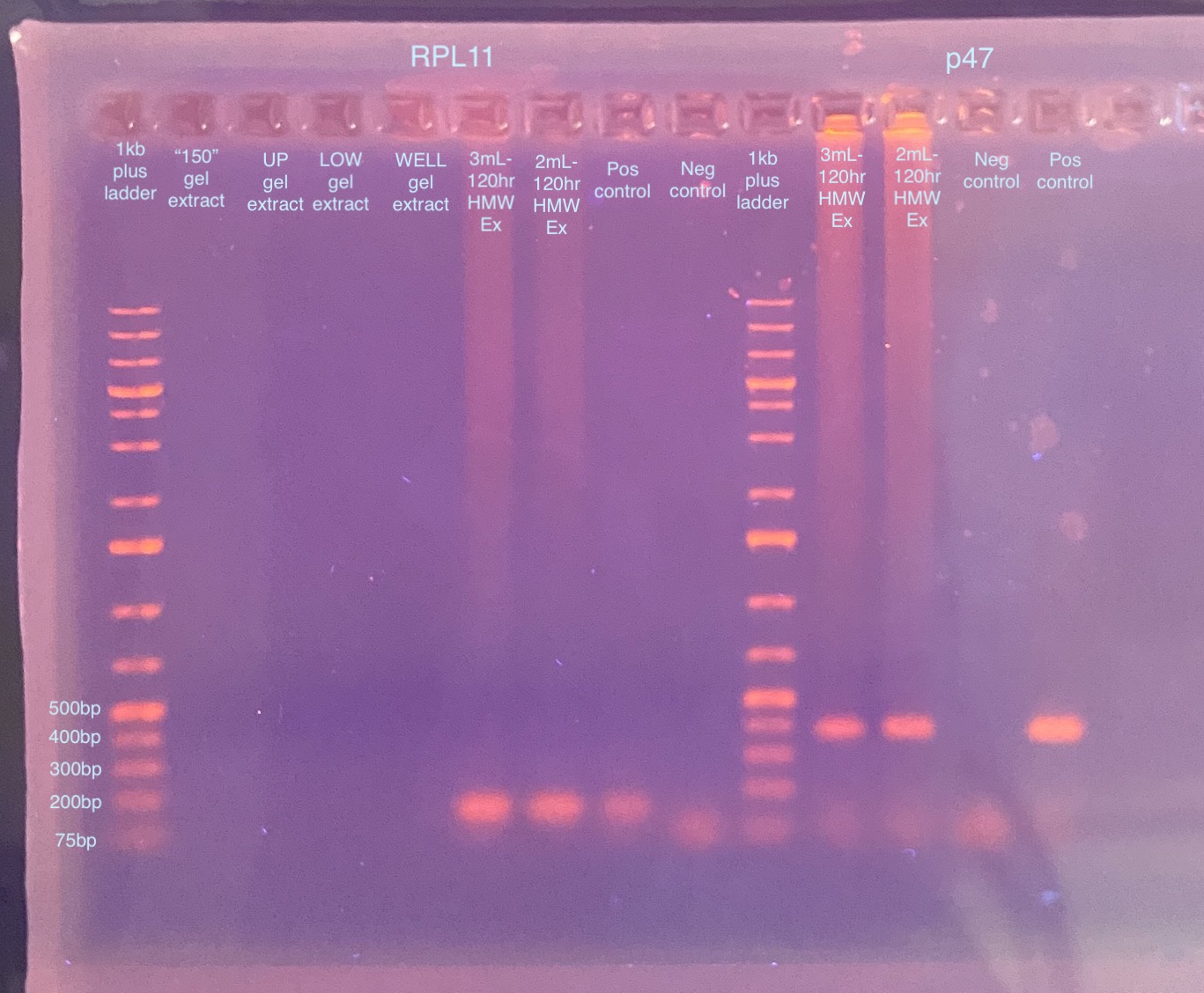
The 3mL-120hr and 2mL-120hr samples are positive for p47! For RPL11, it looks like the fragment should actually only be 150bp in length (info from Rob), so it does seem to have amplified ok for 3mL-120hr and 2mL-120hr. But there is no RPL11 amplification for the gel extraction samples. I will need to redo the gel extraction and try a “dry” spin after the washes to get rid of any residual ethanol (my only idea for why it’s not working atm).