16 Hour Overnight Gel and Zymoclean Large Fragment DNA Recovery Kit on 3mL-120hr HMW DNA Sample
Notes
- Using 3mL-120hr HMW DNA sample (~115ng/ul)
- Skipped wells in the gel when loading to give ample room for slicing
- I also did extractions on a just agarose gel piece as an extraction negative control, and on the PFG marker as an extraction “positive” control
- Minimized time on UV by cutting out the main HMW sample block in 1 go and slicing it into 3
- Zymo kit says it can recovery DNA up to 200kb in length, so I’m hoping it keeps my DNA intact
- I implemented all modifications to the Zymo kit to maximize DNA yield and length:
- Heated elution buffer to 70 degrees C
- Did two elution centrifuges
- Incubated elution liquid on the column for >1 minute each time (5 min each)
20211130 Loading and Running Gel
- 0.74% gel mix:
- 180mL 1X TAE
- 1.3g agarose
- Microwaved for ~4min
- Pipetted 1mL into a 1.5mL tube and saved on the heat block at 65 degrees C make sure the heat block is on and running
- Made sure the large tray had well sealed tape barriers
- Let the gel liquid cool for ~10 min until pouring into the tray and placing in the combs
- Gel cooled in ~20 min or less
- Added 1 round of PFG ladder to the left-most well
- Let the 1mL saved gel cool for ~3 min outside of the heat block
- Filled the PFG well with cooled gel liquid to the top of the well to seal the round in
- Waited ~3 minutes for the wells to cool
- Made up 48kb ladder:
- 1.2ul ladder
- 6ul loading dye
- 28.8ul molecular grade water
- Placed the gel tray into the box after removing the tape
- Mixed 2ul of loading dye with 10ul of 3mL-120hr sample (with a clipped tip) and added it to the 3rd well in the gel
- Added 16ul 48kb ladder to the 5th well in the gel
- Set the timer for 16.5 hours at 40 volts and started it at ~5:06pm
20211201 Gel Slicing and Extraction
- Stopped the gel at 8:48am
- Sliced the gel in half and placed the half with samples in the EtBr bath for 1 hour
- While that was in the bath, I made up final tubes and weighed tubes for the starting weight for the gel extraction
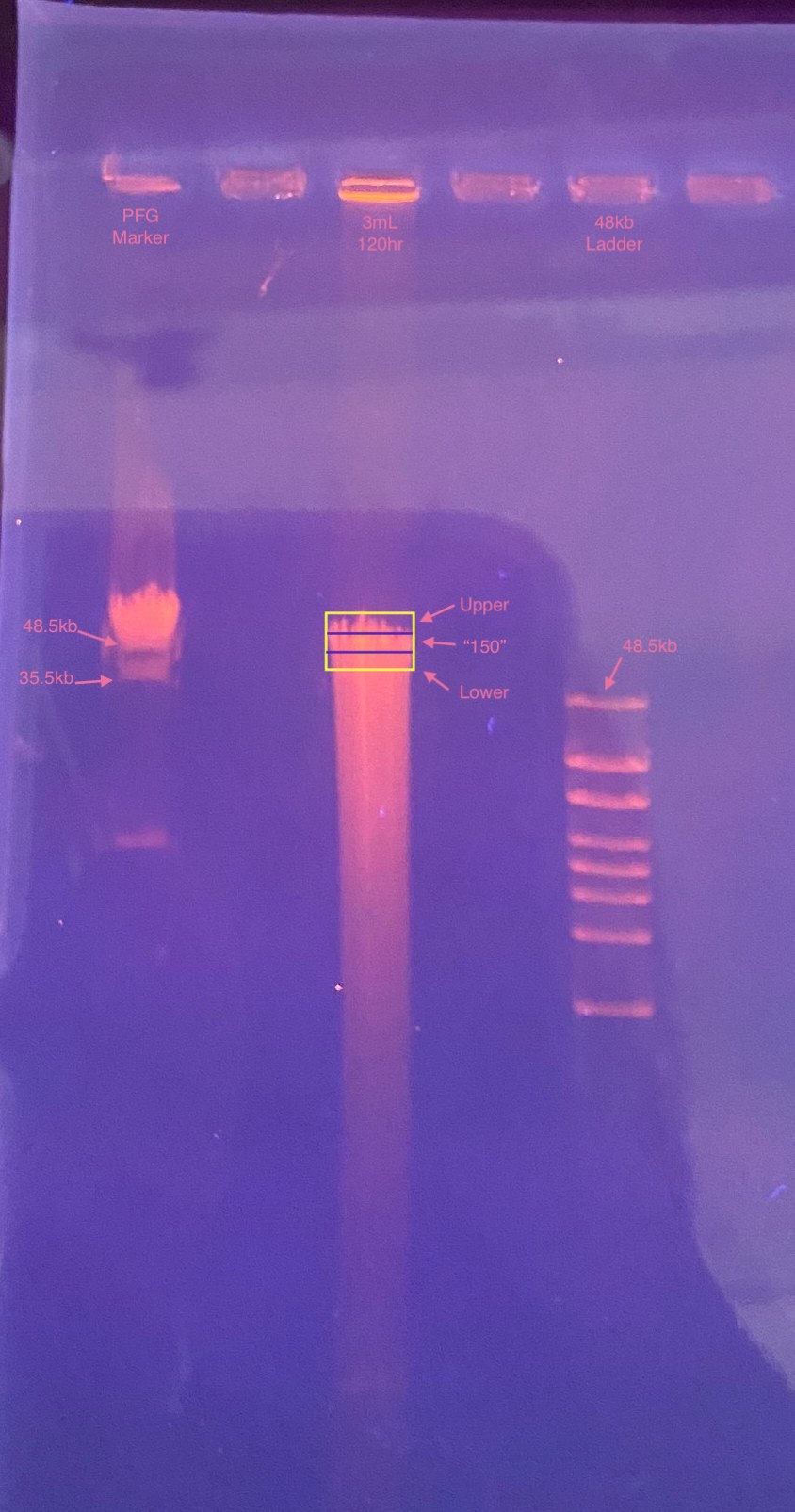
- After 1 hour: put on PPE for looking at UV and took picture of the gel
- Cleaned fresh razor blade with ethanol before use
- Quickly sliced out a square of the whole “HMW” section of the 3mL-120hr sample and of the PFG marker, then kept the UV off for all other slicing
- Sliced the 3mL-102hr piece into 3 for “upper”, “150”, and “lower” sections. This was harder after the square came out of the gel, but it was still possible
- Sliced a section of the gel off to the side where no wells were used for the gel control
- Sliced under the 3mL-120hr sample well
- Gel Slices and distributions:
- Gel slices (no UV)

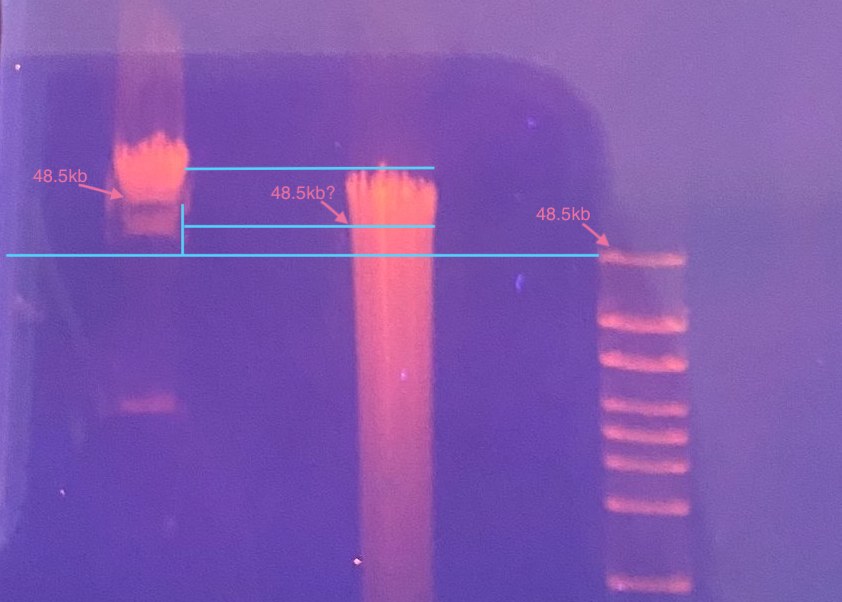
- I also noticed later that my sample seemed maybe not so HMW as previous ones, however the 48kb ladder did not match up with the 48.5kb band in the PFG marker, so I wonder if the gel ran a little slanted?
- Cleaned up and disposed properly all EtBr touching items
- Weighted every sample tube with the gel in it, and calculated the gel weight, and the 3X gel weight
| tube name | tube weight (mg) | tube weight with gel (mg) | gel weight (mg) | 3X gel weight volume (ul) |
|---|---|---|---|---|
| 3mL-150 | 993.5 | 124.9 | 31.4 | 94.2 |
| 3mL-UP | 973.8 | 999.2 | 25.4 | 76.2 |
| 3mL-LOW | 984.4 | 1012.8 | 28.4 | 85.2 |
| 3mL-WELL | 987.5 | 1054.3 | 66.8 | 200.4 |
| Gel-Control | 989.2 | 1068.2 | 79 | 237 |
| PFG-Ladder | 983.4 | 1052.8 | 69.4 | 208.2 |
- New kit: added 24mL 100% ethanol to the DNA wash buffer
- Added 3 volumes of gel weight (5th column in table)of ADB to each sample tube (kit buffer)
- Incubated sample tubes at 50 degrees C in the heat block for 5 minutes (they seemed completely dissolved after this)
- After this, the heat block was bumped to 70 degrees and elution buffer was incubated until later
- Clipped tips of pipettes and added the total volume of each dissolved gel tube to individual spin columns and collection tubes provided by the kit
- Centrifuged spin columns for 1 minute at 12,000rcf
- Pipetted out flow through
- Added 200ul of DNA wash buffer to each column
- Centrifuged columns for 30 seconds at 12,000rcf
- Pipetted out flow through
- Added another 200ul DNA wash buffer to each column
- Centrifuged for 30 seconds at 12,000rcf
- Transferred columns to final 1.5mL tubes
- Added 15ul of 70 degree C DNA elution buffer directly to the filters in the columns
- Incubated the columns on the bench for 5 minutes
- Centrifuged the columns for 30 seconds at 12,000rcf
- Added 15ul of 70 degree C DNA elution buffer directly to the filters in the columns
- Incubated the columns on the bench for 5 minutes
- Centrifuged the columns for 30 seconds at 12,000rcf
- Put the tubes on ice for Qubiting
- Qubit HS DNA Assay:
- 3mL-150: 3.47ng/ul
- 3mL-UP: 2.09ng/ul
- 3mL-LOW: 2.18ng/ul
- 3mL-WELL:1.13ng/ul
- Gel-Control: too low
- PFG-Ladder: 4.85ng/ul