2mL and 3mL Input into the Genomic Tip Blood & Cell Culture DNA Midi Kit HMW Extraction Kit with 120hr DiNV Infected Dv-1 Cells from Kent
Notes
- Both the 2mL and 3mL tubes are from the 120hr: Dv-1 120hr inc-11-2-21 frozen 11-7-21 sample. See previous post for information on the sample
- Going to leave the samples resuspending for a few days, put in the fridge over the weekend, and quantify on Monday
- Used the swing bucket rotor that does 4816g in the Slusky Lab, it worked well last time
- Samples were thawed in the 4 degree fridge overnight prior to use
20211109
Sample Washing
- Placed 1X PBS and molec grade water on ice
- Samples were removed from the fridge (in 1mL aliquots), and divided into 2 15mL tubes. One with 3mL input and one with 2mL input. All pipetting was with clipped pipette tips
- Set centrifuge on the 5th floor to go to 4 degrees
- Added 7mL cold PBS to the tube with the 3mL sample, added 8mL cold PBS to the tube with the 2mL sample
- Centrifuged tubes at 1,500rcf for 10 minutes at 4 degrees C
- Removed supernatant from each tube without disturbing the pellet
- A nice pellet had formed for the 3mL sample, a slightly smaller one for the 2mL sample

- A nice pellet had formed for the 3mL sample, a slightly smaller one for the 2mL sample
- Added 10mL cold PBS to each tube
- Centrifuged tubes at 1,500rcf for 10 minutes at 4 degrees C
- Removed supernatant
- Added 2mL cold PBS to each tube
- Added 2mL cold buffer C1 to each tube
- Added 6mL cold molecular grade water to each tube
- Vortexed to get the pellet to resuspend
- Let tubes sit on ice for 10 minutes
- Centrifuged 1,300rcf for 15 minutes at 4 degrees C
- Removed supernatant without disturbing the pellet
- There was a small pellet in each sample
- Added 1mL cold buffer C1 to each tube
- Added 3mL cold molecular grade water to each tube
- Vortexed breifly
- Centrifuged 1,300rcf for 15 minutes at 4 degrees C
- Removed supernatant
Incubation
- Added 5mL buffer G2 to each tube
- Added 95mL proteinase K to each tube
- Vortexed tubes briefly
- I had to vortex these a lot
- Placed tubes in the incubator at 50 degrees C for ~60 minutes
Genomic Tip Extraction
- Set up two tips over 50mL conicals
- Added 4mL buffer GBT to each tip and let drip
- Drip times were pretty consistent between the tips
- Vortexed sample tubes briefly
- Cut p1000 tips for each samples
- Added total volume (~5mL) of each sample to their respective tips and let drip
- Placed buffer QF in the incubator to warm to 50 degrees C
- Added 7.5mL of buffer QC to each tip
- Transferred tips to a new 15mL waste conical
- Added 7.5mL of buffer QC to each tip
- Transferred tips to new 15mL conicals labeled for final tubes
- Added 5mL of warmed buffer QF to each tip and let drip
Precipitation
- Added 3.8mL of 100% isopropanol to each elutent tube and inverted multiple times to mix (it looked like there was ~5.5mL of volume)
- Centrifuged tubes in the Slusky Lab at 4 degrees C 4,816rcf for 45 min (I increased the time here to hopefully get more DNA to precipitate)
- Afterwards it was very hard to see a pellet but I think there was one
- Put the tubes back on ice and brought to 4055
- Removed the supernatant from each tube, pipetting from the side of the tube without the white patch to avoid any pellet
- Added 2mL cold 70% ethanol to each tube
- Vortexed tubes briefly
- Centrifuged tubes for 15 minutes at 8,816 rcf at 4 degrees C in the same centrifuge
- Tubes were transferred on ice again
- Removed the supernatant from each tube, pipetting from the side of the tube without the white patch to avoid any pellet
- Flipped the tubes upside-down on a kim wipe and let them sit for ~30-40 minutes to dry
- Added 50ul of 1X TE to each tube
- Here I saw what should be the DNA pellet come up when I added the TE
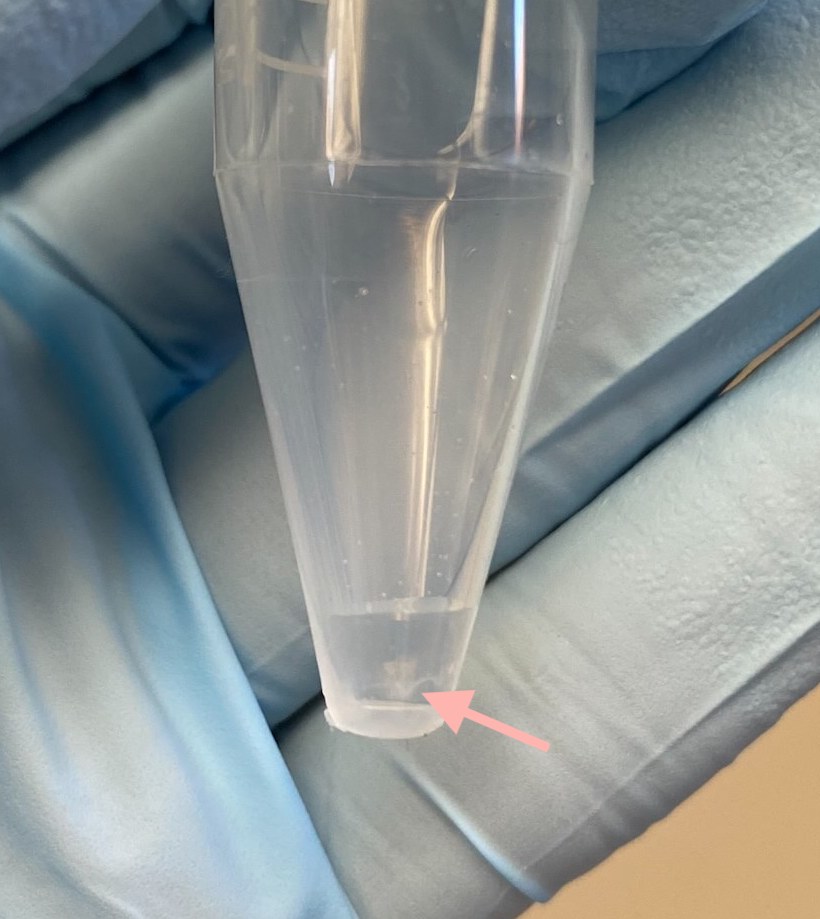
- Here I saw what should be the DNA pellet come up when I added the TE
- Tubes were placed in the incubator upright at 55 degrees for 1 hour
- Tubes were then placed on a rocking machine overnight, and will probably be left until Friday out, and then be put in the fridge for the weekend
20211129 Qubit
- Measured top and bottom of each tube liquid because HMW DNA can be clumpy
- 3mL top: 110ng/ul
- 3mL bottom: 120ng/ul
- 2mL top: 42.3ng/ul
- 2mL bottom: 43.2ng/ul
- I noticed that the DNA might not be completely in suspension. Even after being left to resuspend for days there were still flakes of DNA(?) at the bottom of the tubes. I flicked them many times but they didn’t go into solution. I may have over dried the pellet by leaving it for ~30 minutes (google suggestion), and I will try to pipette out any excess ethanol next time and dry for only 2 minutes.