Methylation Comparison WGBS With Pico Methyl Seq Library Prep Kit First Try
Using the Zymo Pico Methyl Seq Kit on 3 Pocillopora and 3 Montipora samples for the purpose of whole genome bisulfate sequencing to compare to RRBS and MBD-BS preparations
Samples for methylation comparison are from the Holobiont Integration Project, and were extracted by Emma Strand or myself, see her notebook posts for the extraction specifications: 20190805 and 20180823, 20190718 and 20190903.
Started with 100ng of DNA for each sample, and spiked in 5ng of unmethylated lamda DNA (originally from the RRBS kit) to test bisulfite conversion. Then increaesd the volume to 20µl for the start of bisulfite conversion.
Samples are all Soft homogenization DNA, and if elutions were separated they were the second elution.
| Sample | volume DNA (100ng) | Volume spike in | Volume 10mM Tris HCl |
|---|---|---|---|
| 1041 | 1.6 | 2 | 16.4 |
| 1471 | 1.8 | 2 | 16.18 |
| 1637 | 1.15 | 2 | 16.85 |
| 1101 | 3.69 | 2 | 14.31 |
| 1548 | 4.33 | 2 | 13.67 |
| 1628 | 5.88 | 2 | 12.12 |
Bisulfite Conversion
- Added 130µl lightning conversion reagent to each sample in at PCR tube
- Put tubes in the thermocycler Pico bisulfite conversion program
Cleanup
- Added 600µl M-binging buffer each to six spin columns
- Added 150µl of the BS reaction (all) to each individual tube
- Invert columns to mix
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 100µl M-Wash buffer to each column
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 200µl L-desulfonation buffer to each column
- Let them sit for 15 minutes
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 8µl warmed (56C) DNA elution buffer to each column and let sit for 1 minute in new 1.5mL tubes to collect
- Centrifuged columns at 12,000 rcf for 30 seconds
Amplification with PrepAmp Primers
- Made Priming master mix on ice:
- 2µl 5X PrepAmp buffer * 6.2 = 12.4µl
- 1µl PrepAmp Primers (40µM) * 6.2 = 6.2µl
- Made new PCR tubes with 3µl of PrepAmp MM and 7µl of bisulfite treated DNA
- Kept those on ice
- Made PrepAmp Mix on ice:
- 1µl 5X PrepAmp buffer * 6.2 = 6.2µl
- 3.75µl PrepAmp PreMix * 6.2 = 23.25µl
- 0.3µl PrepAmp polymerase * 6.2 = 1.86µl
- Set thermocylcer program with lid temp restricted to 25 degrees C and place samples inside and run:
- 98 for 2 minutes
- 8 degrees for 1 minute
- 8 degree hold
- During hold vortex, spin tubes down, add 5.05µl PrepAmp Mix to each tube, vortex, spin down, and place back in thermocycler
- 8 degrees for 4 minutes
- 16 degrees for 1 minute with 3% ramp rate
- 22 degrees for 1 minute with 3% ramp rate
- 28 degrees for 1 minute with 3% ramp rate
- 36 degrees for 1 minute with 3% ramp rate
- 36.5 degrees for 1 minute with 3% ramp rate
- 37 degrees for 8 minutes
- repeat back from the first step one time through again
- During hold, vortex, spin tubes down, tried to add 0.3µl PrepAmp Polymerase to each tube, it wouldn’t really come out of the tip so I ended up adding .5µl, vortex, spin down, and place back into thermocycler
Cleanup with DNA Clean and Concentrator Columns (DCC)
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 107.45µl of DNA binding buffer
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (15.35µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 12µl warmed elution buffer to each column directly
- Incubated 1 minute
- Centrifuged 12,000 rcf 30 seconds
First Amplification
- Made 1st Amp master mix:
- 12.5µl 2X Library Amp Mix * 4.2 = 52.5µl
- 1µl Library Amp Primer(10µM) * 4.2 = 4.2µl
- Added 13.5µl MM to new PCR tubes
- Added 11.5µl of cleaned and concentrated DNA sample to the appropriate new PCR tube note here, samples 2 and 6 had a lot more than 11.5µl in them, could have been carryover of wash buffer in the column, only 11.5µl were used for this step though to keep volumes the same. What was left over I froze in the -20
- Vortexed, spun down, and placed in thermocycler program 1st Pico Methyl Amp program 6 cycles for the 100ng input
Cleanup with DCC
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 175µl of DNA binding buffer
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (25µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 12.5µl warmed elution buffer to each column directly
- Incubated 1 minute
- Centrifuged 12,000 rcf 30 seconds
Amplification with Index Primers
- Made new PCR tubes for each sample with 12.5µl Library Amp Mix
- Index primers are the dual index UDI i5 and i7 indexes. That means each sample needs to get primers from 2 tubes, but the total volume of primers needs to be .5µl (so .25µl each) which is already a lot smaller than what can be easily pipetted. Because of previous issues pipetting .3µl, I made separate tubes with 1µl of each primer pair and then added .5µl of that pre-made pair to each sample
-
Added indexed primers:
sample index pair 1041 i5 1 and i7 1 1471 i5 2 and i7 2 1637 i5 3 and i7 3 1101 i5 10 and i7 10 1548 i5 11 and i7 11 1628 i5 12 and i7 12 - Added 12µl of sample to the appropriate tube (all of the flowthrough from above DCC)
- Vortexed, spun down, and placed in thermocycler program 2nd Pico Methyl Amp program
Cleanup with DCC
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 175µl of DNA binding buffer
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (25µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 12µl warmed elution buffer to each column directly
- Incubated 1 minute
- Centrifuged 12,000 rcf 30 seconds
D5000 TapeStation
See the full report here
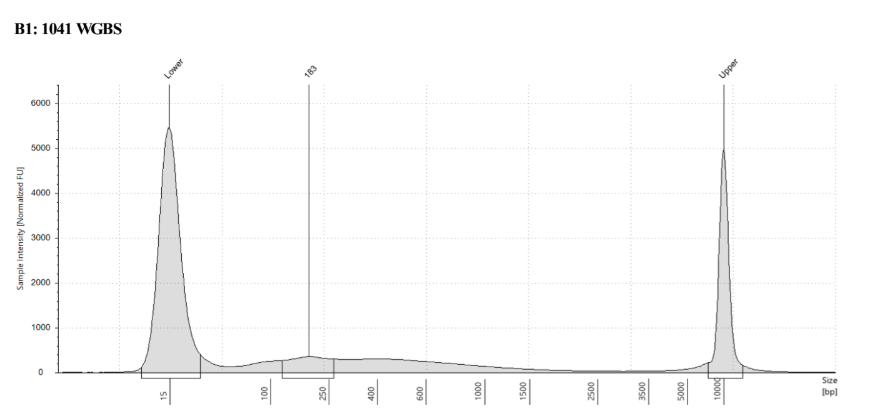
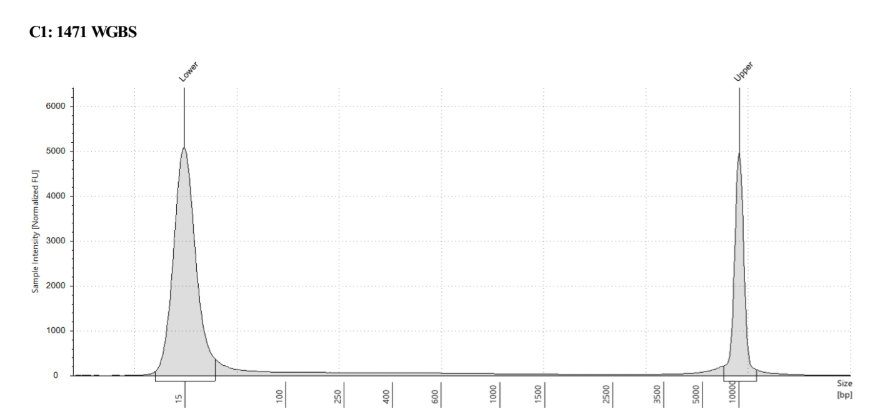
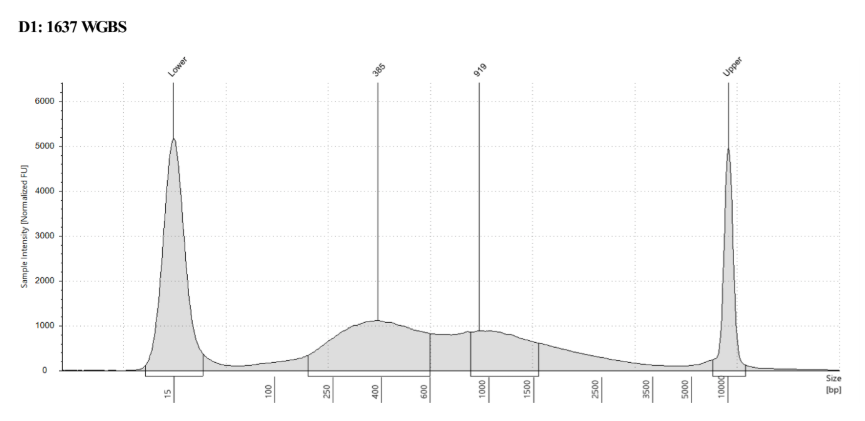
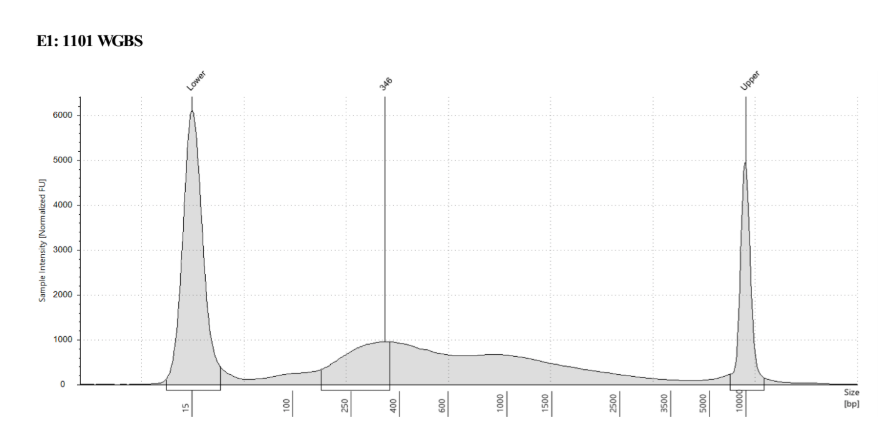
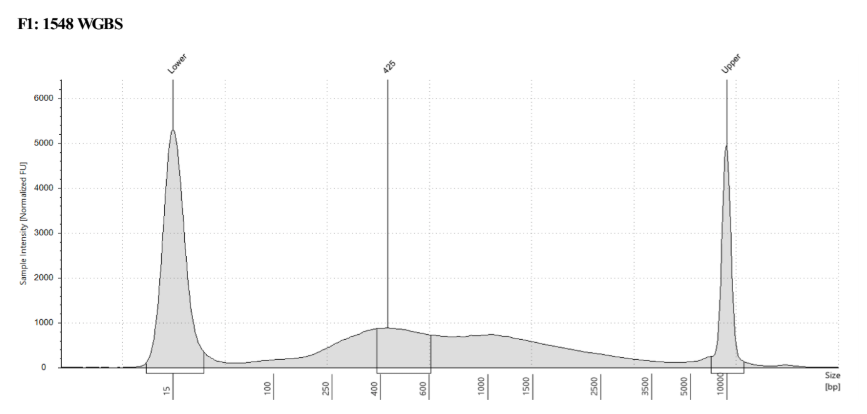
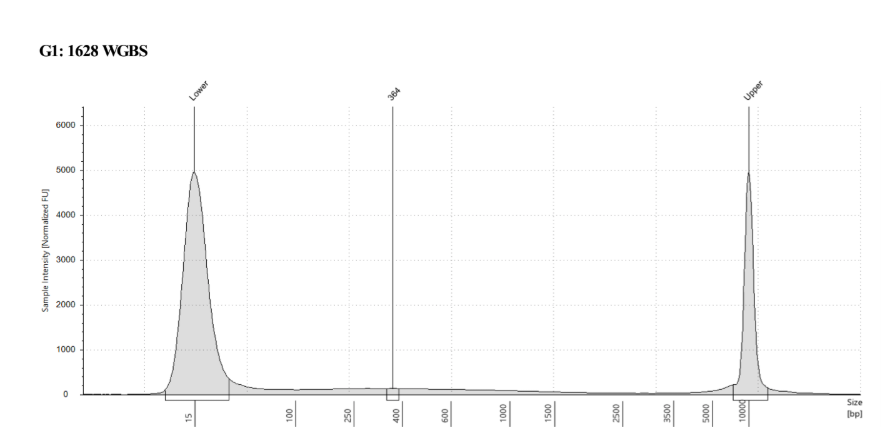
Here, 2 and 6 didn’t work. Which might be because of the extra volume after the DCC before the first amplification. In this post which was a test of this kit, one of the samples had extra volume after a cleanup step and then did not have anything in the sample afterwards either. My guess with what is going on with the other ones is that there was too much input, these traces look similar to 221 NC in that same above post, where I had also started with 100ng input.