Pico Methyl Seq Library Prep Kit Test
Testing Zymo Pico Methyl Seq Kit On Bisulfite Converted MBD Enriched Acropora Sperm Samples from Previous Post
Note: using version 1.2 of the kit manual
Amplification with PrepAmp Primers
- Made Priming master mix on ice:
- 2µl 5X PrepAmp buffer * 4.2 = 8.4µl
- 1µl PrepAmp Primers (40µM) * 4.2 = 4.2µl
- Made new PCR tubes with 3µl of PrepAmp MM and 7µl of bisulfite treated DNA (221 and 1431 captured and not captured)
- Kept those on ice
- Made PrepAmp Mix on ice:
- 1µl 5X PrepAmp buffer * 4.2 = 4.2µl
- 3.75µl PrepAmp PreMix * 4.2 = 15.75µl
- 0.3µl PrepAmp polymerase * 4.2 = 1.26µl
- Set thermocylcer program with lid temp restricted to 25 degrees C and place samples inside and run:
- 98 for 2 minutes
- 8 degrees for 1 minute
- 8 degree hold
- During hold vortex, spin tubes down, add 5.05µl PrepAmp Mix to each tube, vortex, spin down, and place back in thermocycler
- 8 degrees for 4 minutes
- 16 degrees for 1 minute with 3% ramp rate
- 22 degrees for 1 minute with 3% ramp rate
- 28 degrees for 1 minute with 3% ramp rate
- 36 degrees for 1 minute with 3% ramp rate
- 36.5 degrees for 1 minute with 3% ramp rate
- 37 degrees for 8 minutes
- repeat back from the first step one time through again
- During hold, vortex, spin tubes down, add 0.3µl PrepAmp Polymerase to each tube, vortex, spin down, and place back into thermocycler
Cleanup with DNA Clean and Concentrator Columns (DCC)
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 105.49µl of DNA binding buffer
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (15.07µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 12µl warmed elution buffer to each column directly
- Incubated 1 minute
- Centrifuged 12,000 rcf 30 seconds
First Amplification (Adapter addition?)
- Made 1st Amp master mix:
- 12.5µl 2X Library Amp Mix * 4.2 = 52.5µl
- 1µl Library Amp Primer(10µM) * 4.2 = 4.2µl
- Added 13.5µl MM to new PCR tubes
- Added 11.5µl of cleaned and concentrated DNA sample to the appropriate new PCR tube
- Vortexed, spun down, and placed in thermocycler program 1st Pico Methyl Amp program
Cleanup with DCC
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 175µl of DNA binding buffer
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (25µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 12.5µl warmed elution buffer to each column directly
- Incubated 1 minute
- Centrifuged 12,000 rcf 30 seconds
Amplification with Index Primers
- Made new PCR tubes for each sample with 12.5µl Library Amp Mix
- Added indexed primers:
- 221 Captured got 0.5µl Index A
- 1431 Captured got 0.5µl Index B
- 221 Not-Captured got 0.5µl Index C
- 1431 Not-Captured got 0.5µl Index D
- Added 12µl of sample to the appropriate tube (all of the flowthrough from above DCC) note, 1431 captured had more that 12µl in the tube, potentially wash buffer that did not full come out? All of that sample was added to the tube for amplification.
- Vortexed, spun down, and placed in theremocycler program 2nd Pico Methyl Amp program
Cleanup with DCC
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 175µl of DNA binding buffer
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (25µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 12µl warmed elution buffer to each column directly
- Incubated 1 minute
- Centrifuged 12,000 rcf 30 seconds
Slightly more than 12µl seemed to come out of 221 not-captured, more wash buffer carryover?
D5000 TapeStation
- Protocol for D5000 tapestation followed exactly
- Sample 1431 Captured did not show anything in the tapestation, see full results)
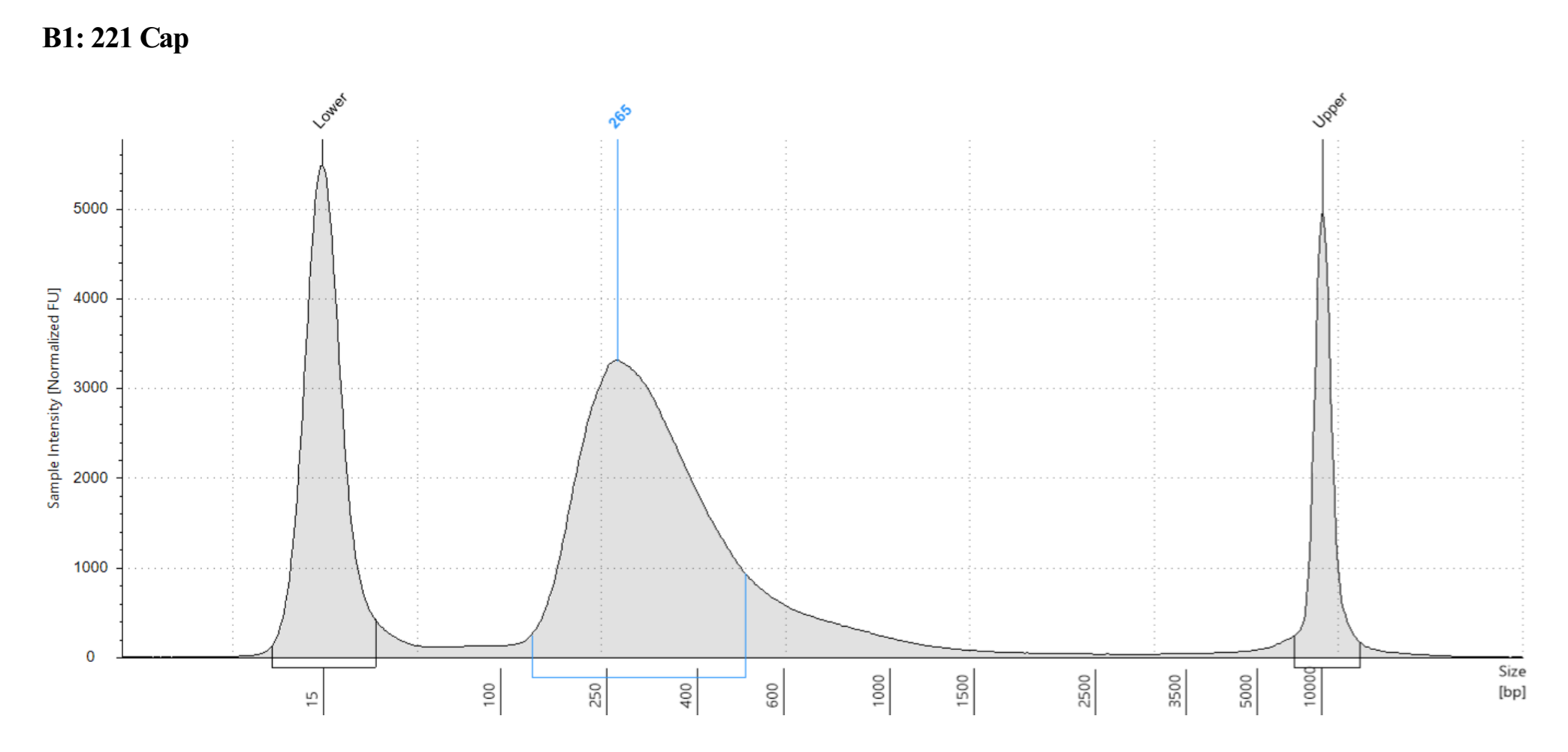
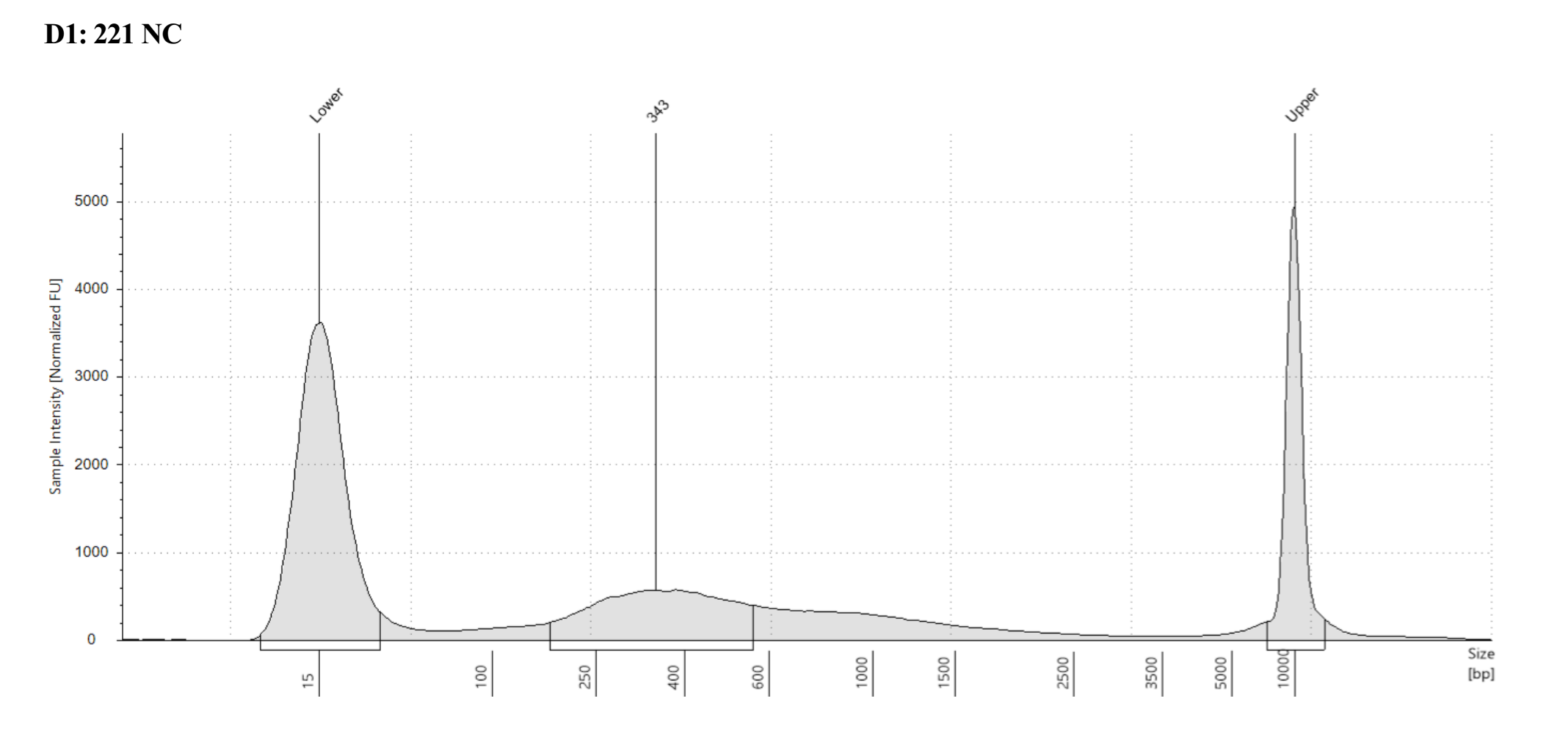
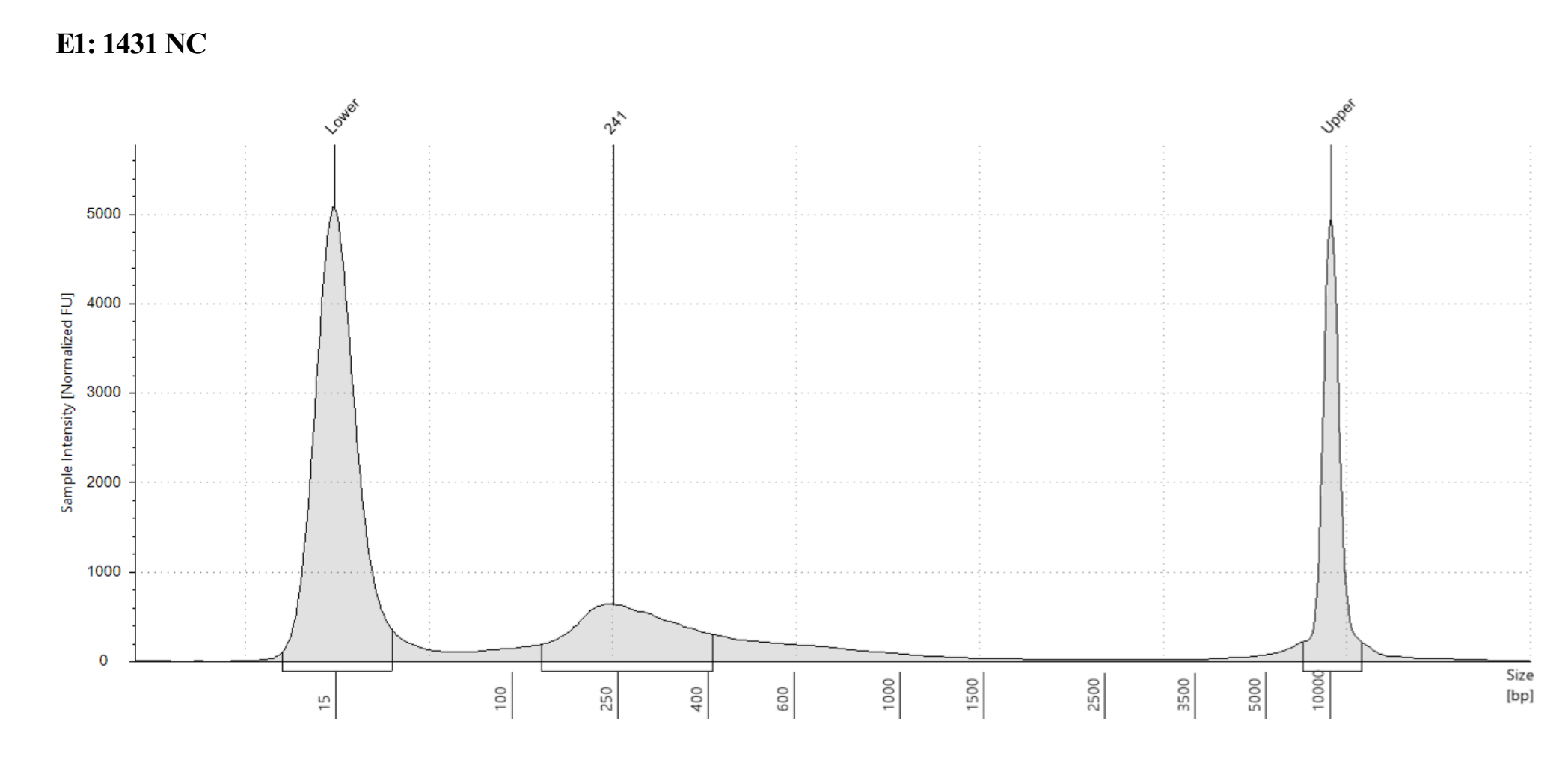
Each one looks different, where potentially 221 captured looks the most like what I think of when I think library, but the others look more like the example in the kit manual. However 221 not-captured does have a long shoulder of larger DNA that was not present in any previous tapestations of that sample….
Written on August 13, 2019