Mo'orea Test Extractions Part 3
Testing to get good RNA out of the Pocillopora and Porites Samples stored in RNA/RNA Shield from Mo’orea
Homogenizing with beads last time may have caused trouble last time because it is hard to get all of the liquid out of the tube with the beads. We have sieves that fit into 1.5mL tubes that would separate the beads from the liquid. The goal of this test was to see if using the sieves gets good RNA and DNA from 4 more coral samples. I took samples from site 3 as to not take too many samples from one site.
Using the Zymo Duet DNA/RNA Extraction Kit
Sample Prep
| Sample # | Site # | Date Collected | Type |
|---|---|---|---|
| 231 | 3 | 2018/03/12 | Massive Porites |
| 216 | 3 | 2018/03/12 | Massive Porites |
| 234 | 3 | 2018/03/12 | Pocillopora verrucosa |
| 238 | 3 | 2018/03/12 | Pocillopora verrucosa |
- Beads from bead tubes were poured into the sample tubes
- Samples were homogenized by vortexing for ~30-60 seconds
- Sieves/strainers were placed in new labeled 1.5mL tubes
- About 1/3 of the sample with the beads was poured into sieve. Not all of the sample fits into the sieve at once, so I poured slowly and only did one species at a time
- Sieves and 1.5mL tubes were centrifuged briefly (pressed start on the centrifuge but did not let it go longer than 10 seconds because the centrifuge couldn’t have the lid put on with the sieves)
- The liquid sample went through into the tube but most of the beads stayed in the sieve
- Repeated pouring the rest of the sample into the sieve and centrifuging quickly. I was not able to get all of the beads out of the tube, even after squirting in a little new DNA/RNA Shield to aid with pipetting up the beads. Tubes with the small amount of beads left were saved in the -20
- After all the sample possible went through the sieve, the sieves with beads and any un-homogenized tissue were labeled with the sample number, parafilmed on the top, and saved in the -20
- There was about 600µl of sample in each tube
- Appropriate volumes of PK Digestion Buffer and Pro K were added
- 60µl PK digestion buffer
- 30µl Pro K
-
- Samples were vortexed, spun down, and placed in the Thermomixer at 55 degrees C for 5 hours (max time recommended in the kit) shaking at 1000rpm
- The rest of the extraction protocol followed the [previous test](last time
Qubit
- Broad Range dsDNA and RNA Qubit protocol
- DNA Qubit was preformed by Marygrace Trusdell
- All samples were read twice
| Sample | DNA Standard 1 (RFU) | DNA Standard 2 (RFU) | DNA 1 (ng/µl) | DNA 2 (ng/µl) | Average DNA | RNA Standard 1 (RFU) | RNA Standard 2 (RFU) | RNA 1 (ng/µl) | RNA 2 (ng/ul) | Average RNA |
|---|---|---|---|---|---|---|---|---|---|---|
| 231 | 208.58 | 23202 | 8.20 | 8.32 | 8.26 | 465 | 11017 | 18.4 | 17.8 | 18.1 |
| 216 | 208.58 | 23202 | 16.2 | 16.1 | 16.15 | 465 | 11017 | 29 | 28.8 | 28.9 |
| 234 | 208.58 | 23202 | 45.4 | 46.4 | 45.9 | 465 | 11017 | 60 | 59 | 59.5 |
| 238 | 208.58 | 23202 | 98.4 | 99 | 98.7 | 465 | 11017 | 146 | 144 | 145 |
TapeStation
- Followed RNA TapeStation protocol
- TapeStation was done with some Mytilus samples extracted by Amy Zyck
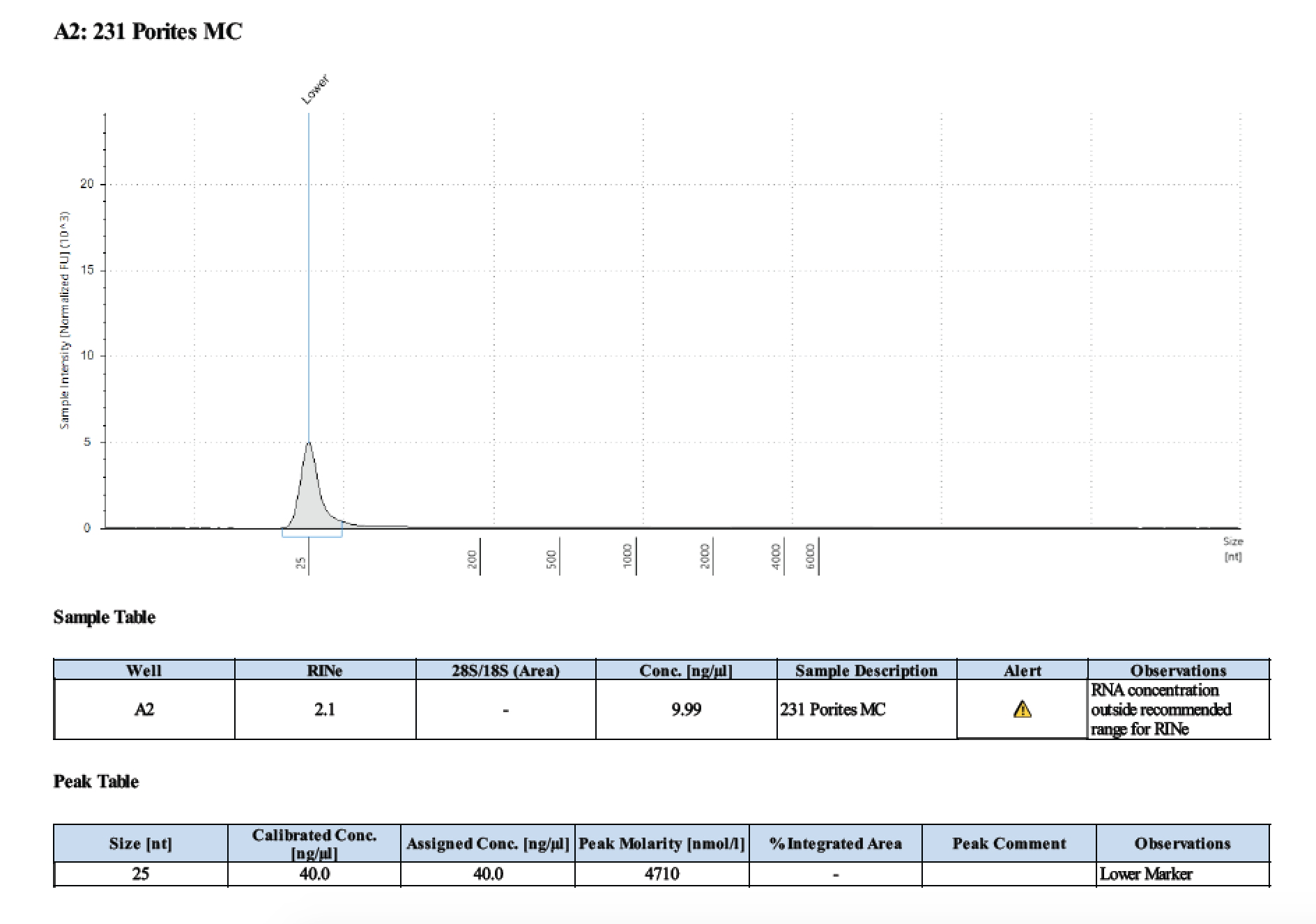
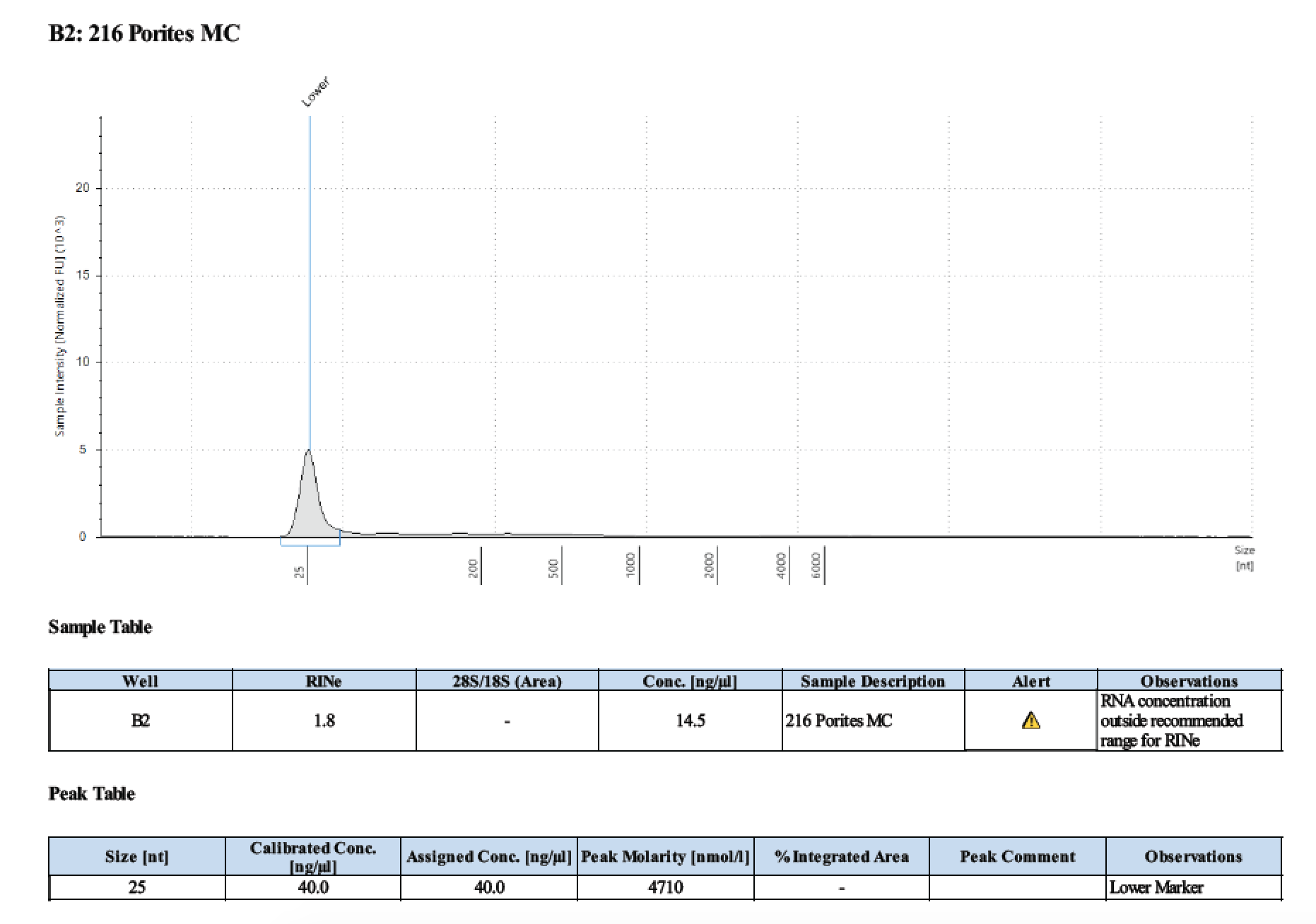
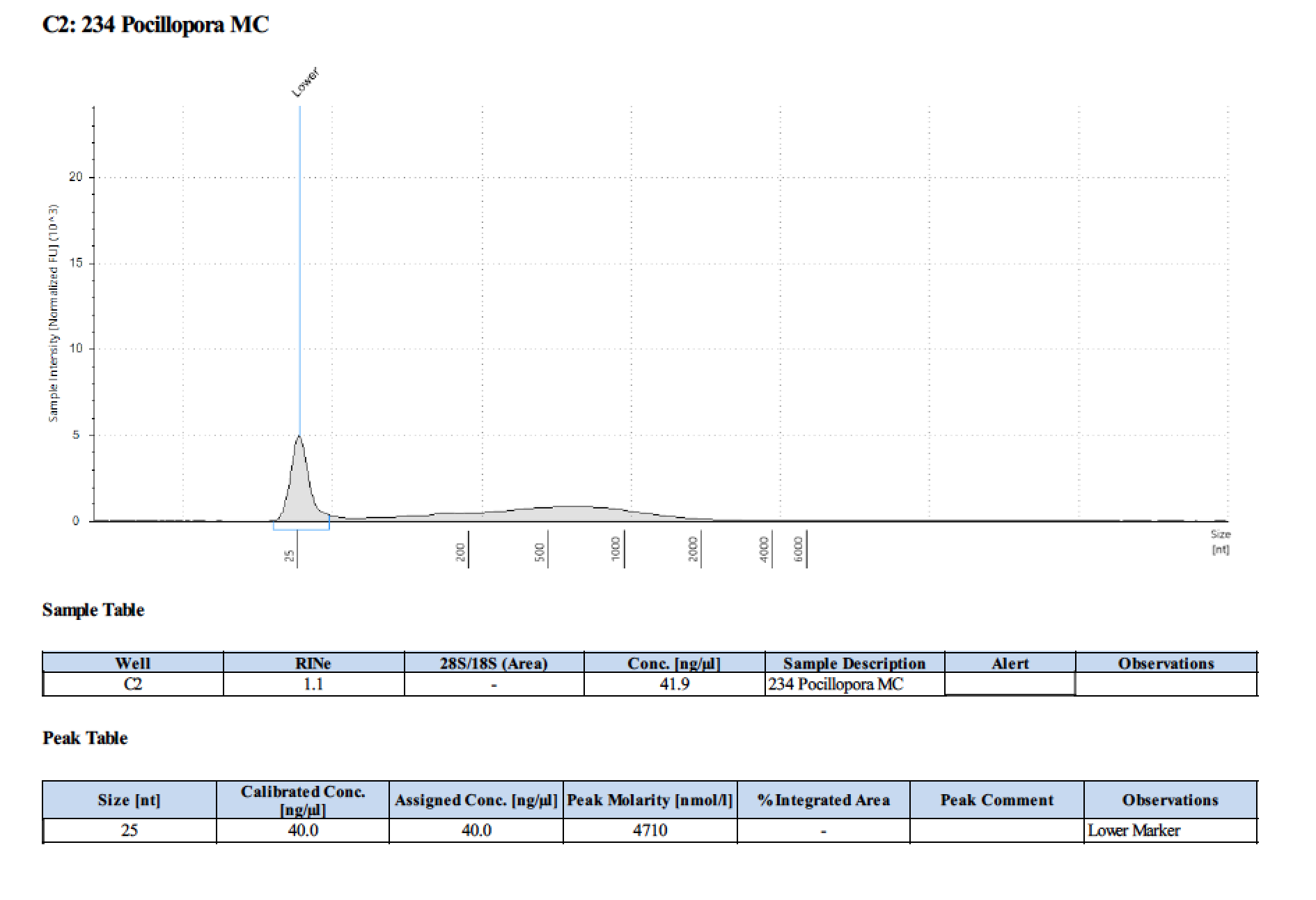
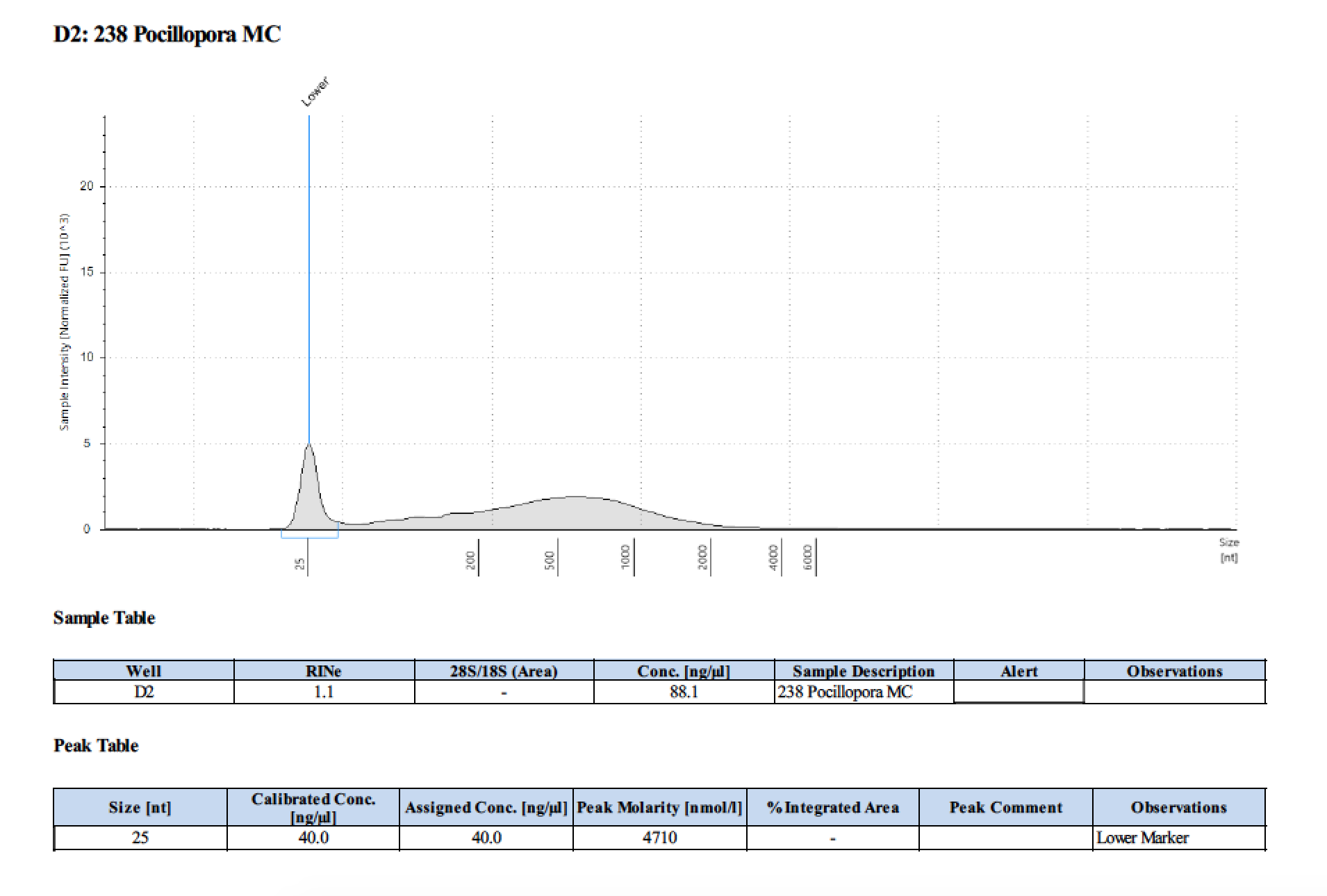
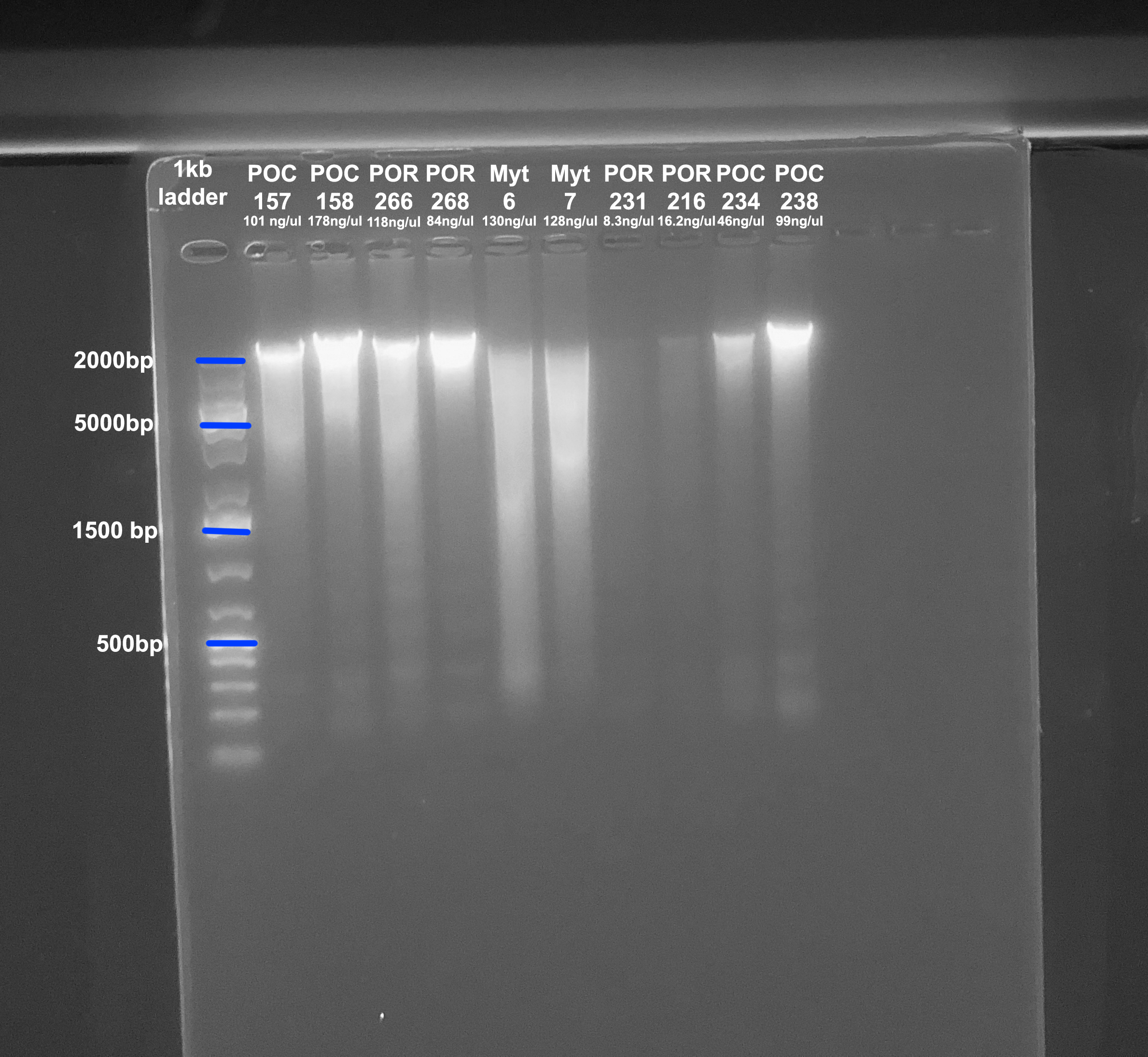
Gel
- A 1.5% agarose gel was ran to check the integrity of the genomic DNA
- Following the PPP Lab protocol
- The first 6 samples are from the next extraction, the last 4 samples are from this extraction
- Below the sample number is the amount of DNA in ng/µl from the Qubit
Written on March 28, 2019