Quality Check of RNA Samples From Biomineralization Project
Purity and Integrity Check of RNA Samples from the Biomineralization Project That Were Extracted in 2019 by Erin Chille
Goal: Re-TapeStation some samples after ~2 years in the freezer, and run them on the NanoDrop in the GSC to get 260/280 and 260/230 ratios. This is to see if the quality of RNA extracted from larvae/eggs is good/minimal contamination. If there is contamination, we may need to modify wash steps in the protocol for up coming RNA extractions for larvae.
Results: TapeStations are surprisingly consistent, and purity is mostly good!
Major Take Aways: Seems like we might not have contamination problems with RNA from larvae. Although I will likely have to use a different extraction procedure than what Erin used in 2019, because Zymo no longer recommends using a heated digestion step. In theory that shouldn’t effect contaminants that could come out with the RNA. I should do the NanoDrop on some of the new samples when I extract.
Sample Information
- I chose 5 samples from the Biomin project that represented a range of RNA concentrations and sampling timepoints (age)
- Extractions were done by Erin Chille: here, here, here, and here
| Sample ID | Extraction # | Extraction Date | Timepoint | Concentration (ng/ul) |
|---|---|---|---|---|
| 183 | 66 | 20190517 | 12:00 | 52 |
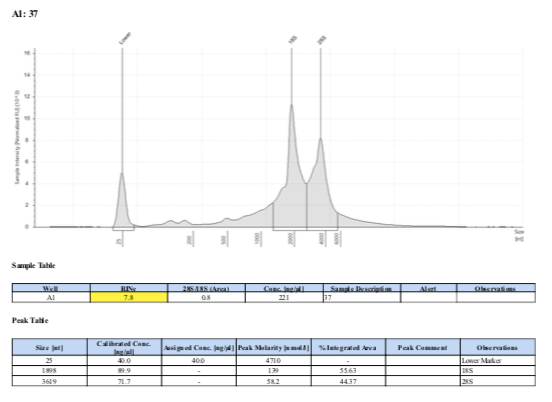
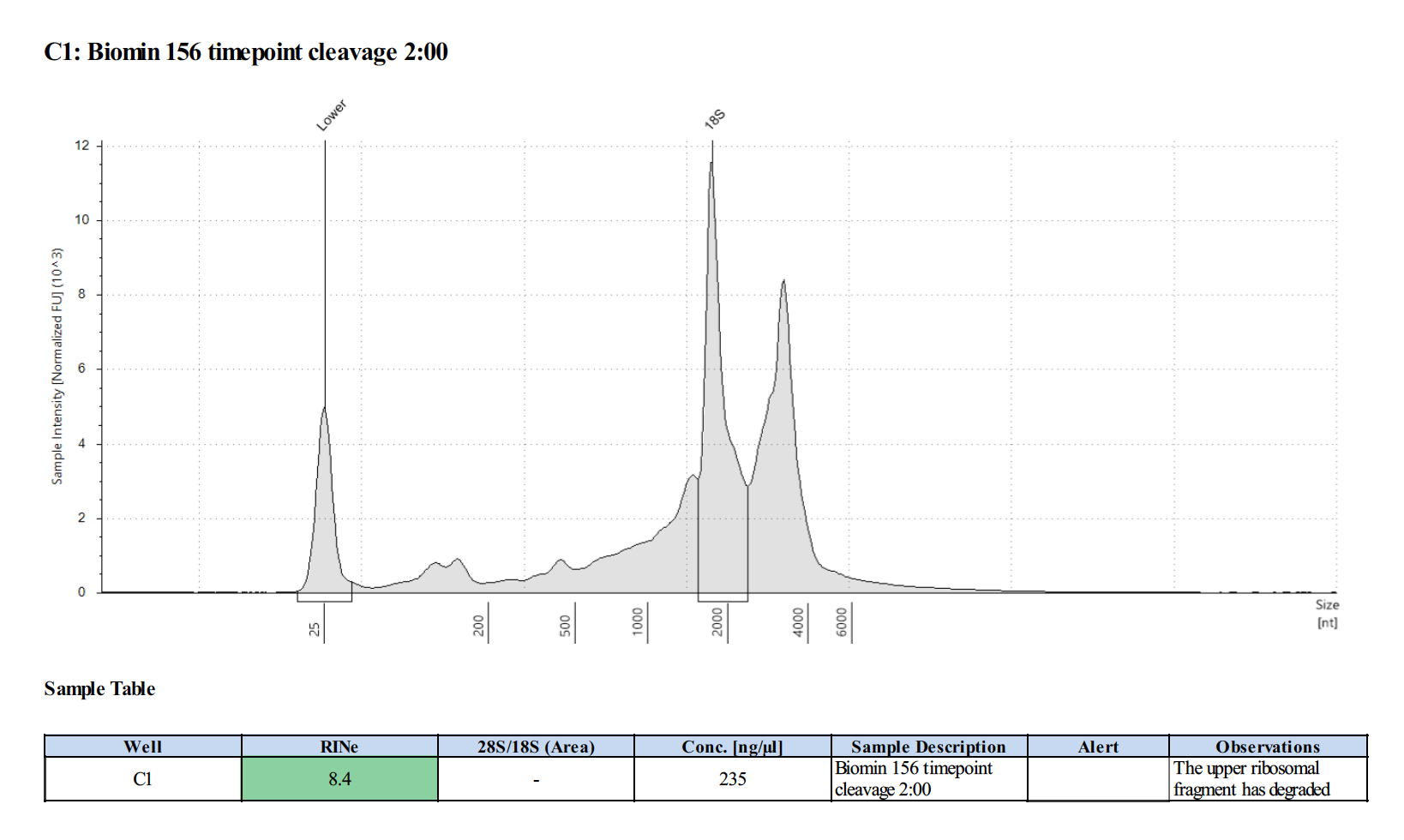
| 156 | 37 | 20190427 | cleavage 2:00 | 266 |
| 129 | 53 | 20190506 | fertilized embryo | 38.5 |
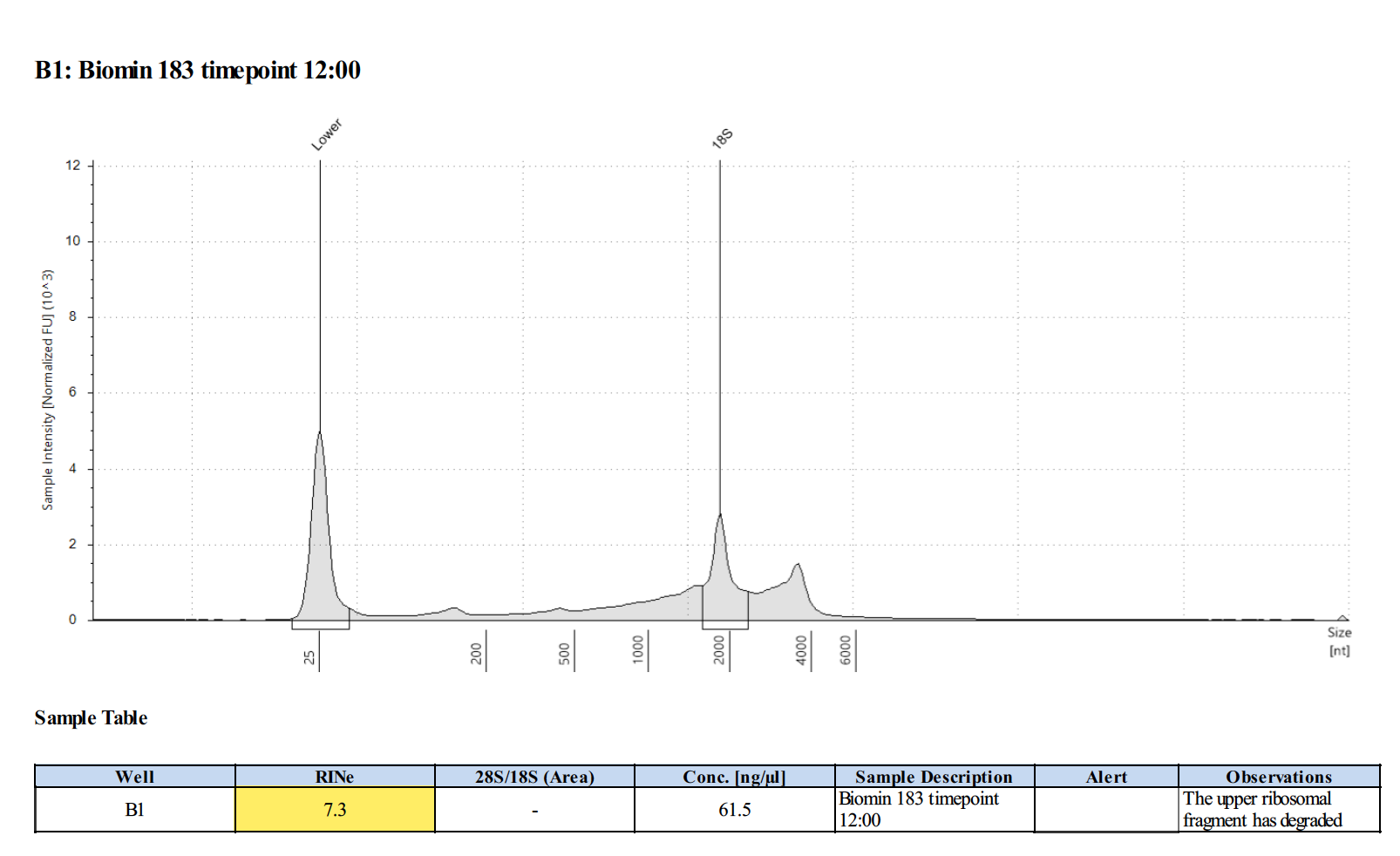
| 164 | 40 | 20190427 | 7:00 | 129 |
| 371 | 25 | 20190322 | 12:00 before settlement | 59.4 |
TapeStation
- TapeStationed the 5 samples to see if quality is similar after ~2 year in the freezer
- TapeStation RNA Protocol
- Results Link
- Comparisons of traces per sample (note samples from 2019 are labeled by the extraction number, see table above):
183 20190517
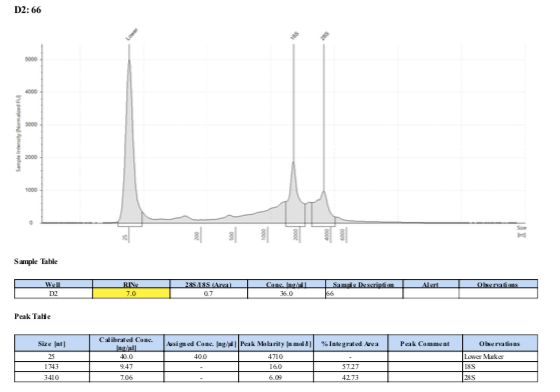
183 20210210
156 20190427
156 20210210
129 20190506
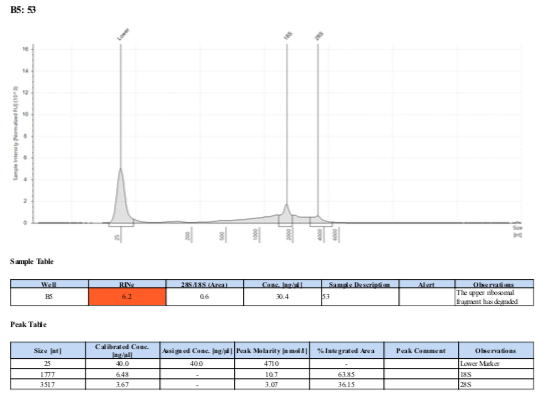
129 20210210
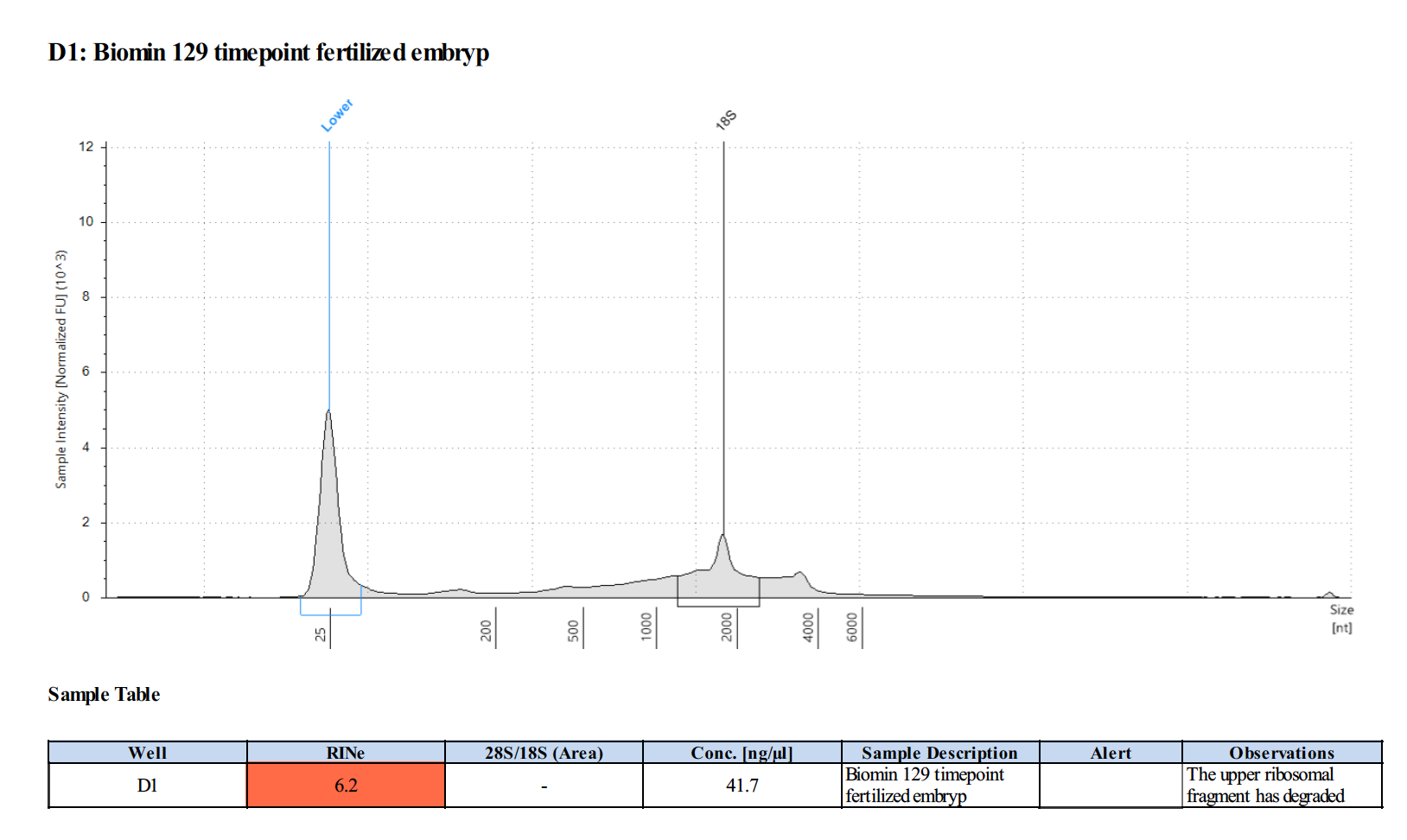
164 20190427
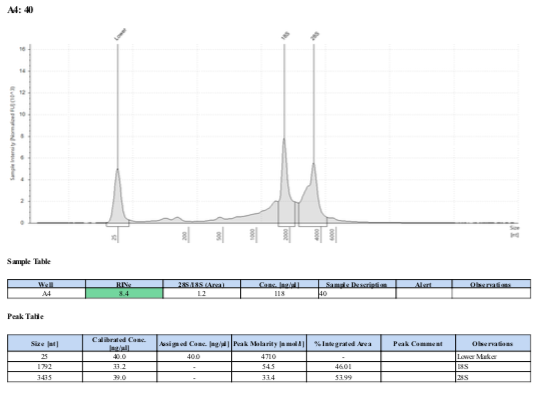
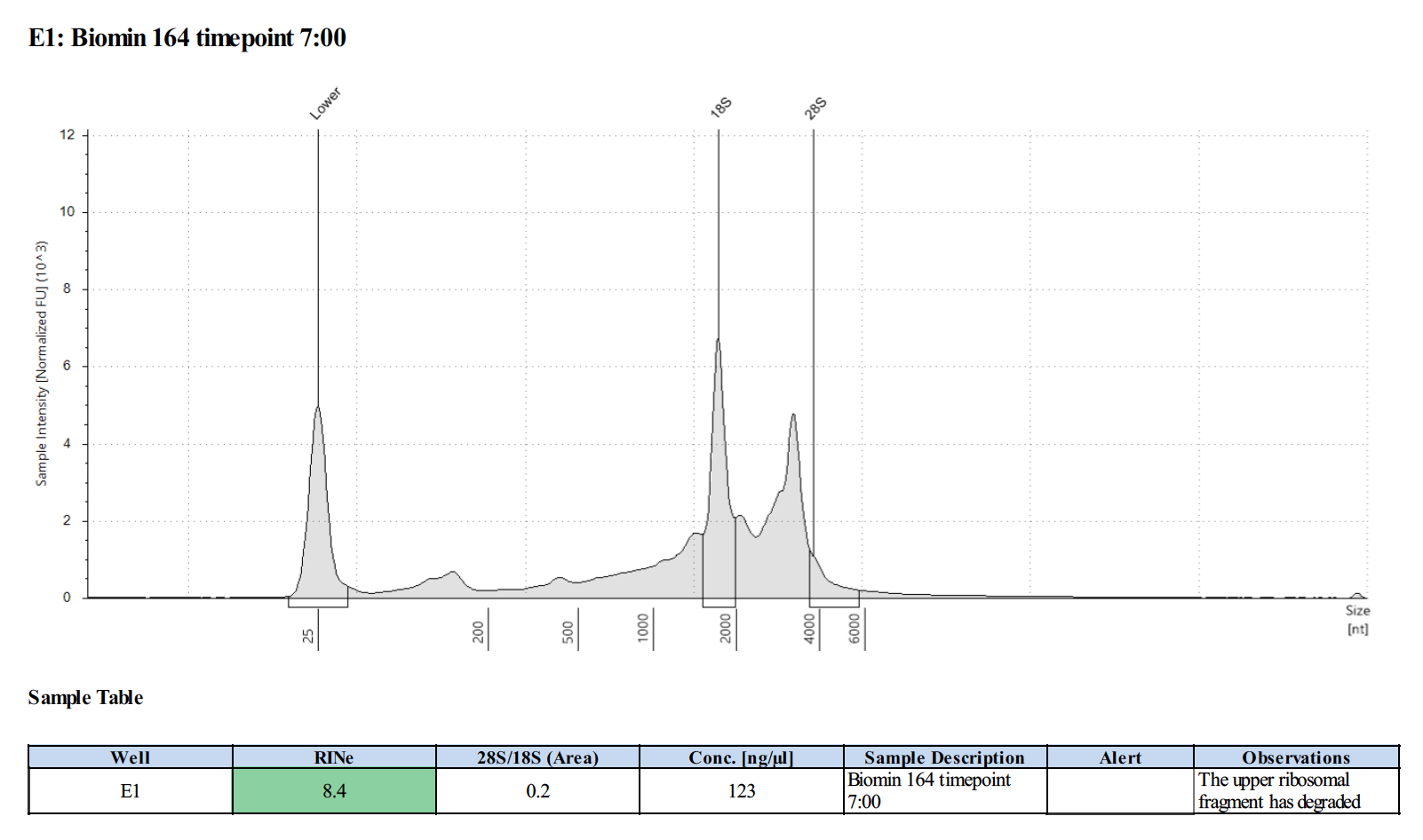
164 20210210
371 20190322
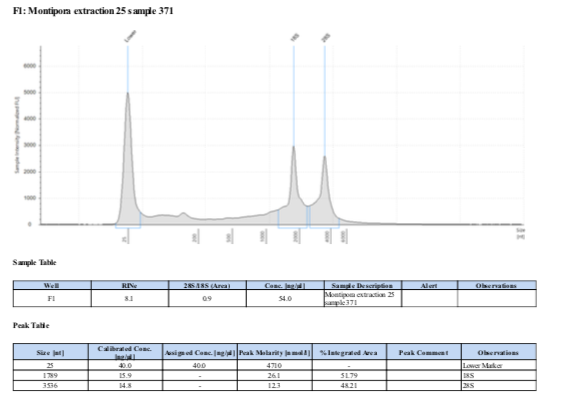
371 20210210

Surprisingly, after 2 years in the freezer the TapeStation traces are almost identical. There seems to be very little degradation over that time. There were 2 freeze-thaws between these dates (once today and once for aliquots for sequencing).
NanoDrop
- Used the NanoDrop 8000 in the URI GSC
- Interpreting RNA NanoDrop Results
- Used the same samples as above, and used the UltraPure water from the RNA bench as the blank and measurement calibration
- Basic steps are as follows (detailed protocol coming soon):
- Took samples, a 1.5mL of the water, a pipette, and filter tips up to the GSC
- Added 2ul of water to the A position and closed “lid” for measurement calibration
- Wiped with kimwipe when done
- Added another 2ul of water to the A position for the blank (water is what RNA is eluted in) and closed lid
- Wiped with kimwipe when done
- Named sample in software with sample ID
- Added 2ul of sample 183 to the A position and closed lid, clicked measure
- Recorded values
- Wiped with kimwipe when done
- Repeated above 4 steps for each sample
- Values:
| Sample | Qubit concentration 2019 | NanoDrop Concentration 2021 | 260/280 | 260/230 |
|---|---|---|---|---|
| 183 | 52 ng/ul | 68.42 ng/ul | 2.06 | 1.77 |
| 156 | 266 ng/ul | 325.8 ng/ul | 2.13 | 2.07 |
| 129 | 38.5 ng/ul | 43.2 ng/ul | 2.12 | 2.05 |
| 164 | 129 ng/ul | 156.9 ng/ul | 2.12 | 2.08 |
| 371 | 59.4 ng/ul | 107.3 ng/ul | 2.10 | 1.66 |
Composite Figure
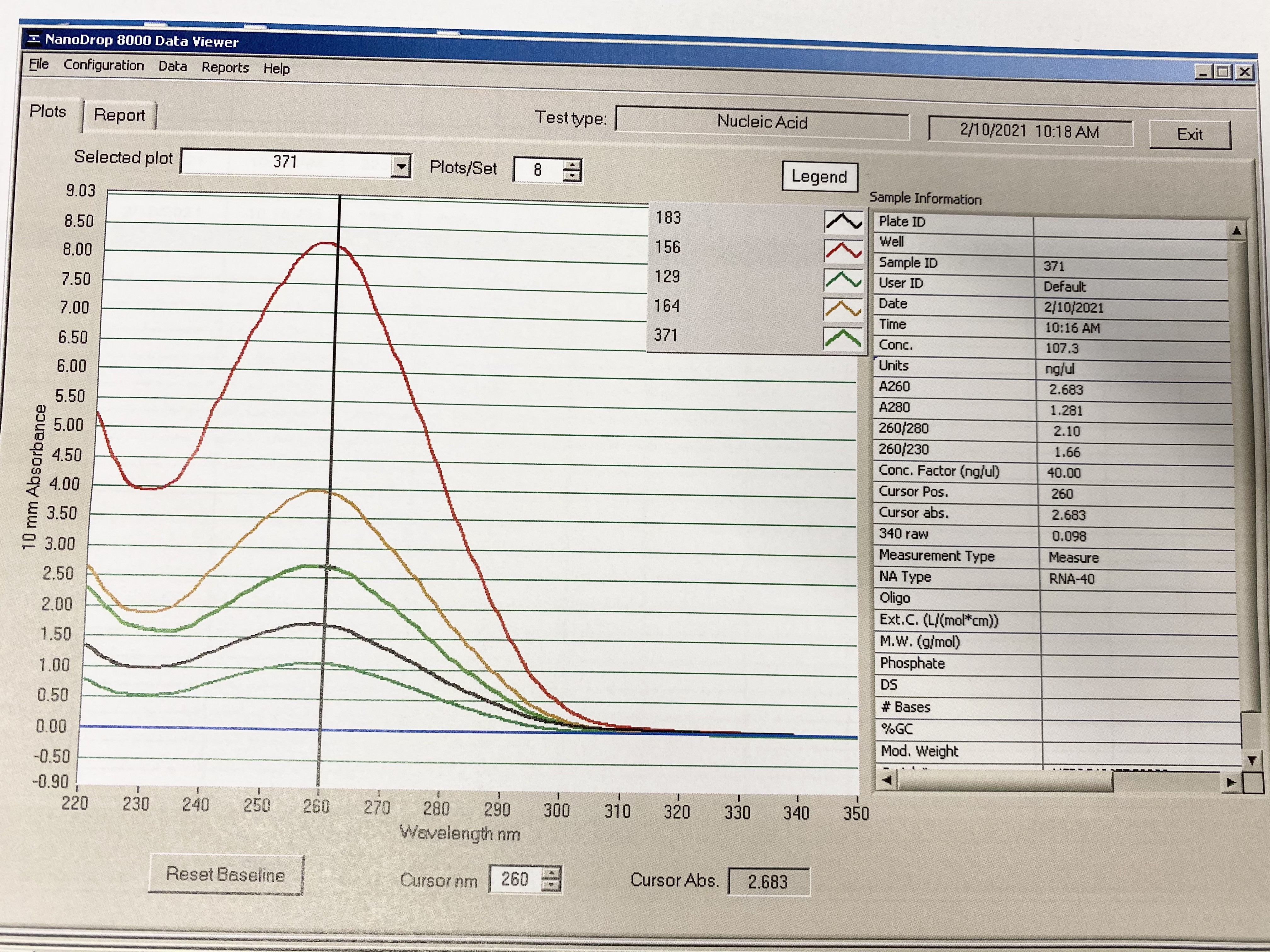 The difference in heights is based on concentration, there was a wide range between samples. All show a pretty good peak and trough.
The difference in heights is based on concentration, there was a wide range between samples. All show a pretty good peak and trough.
Output from Program
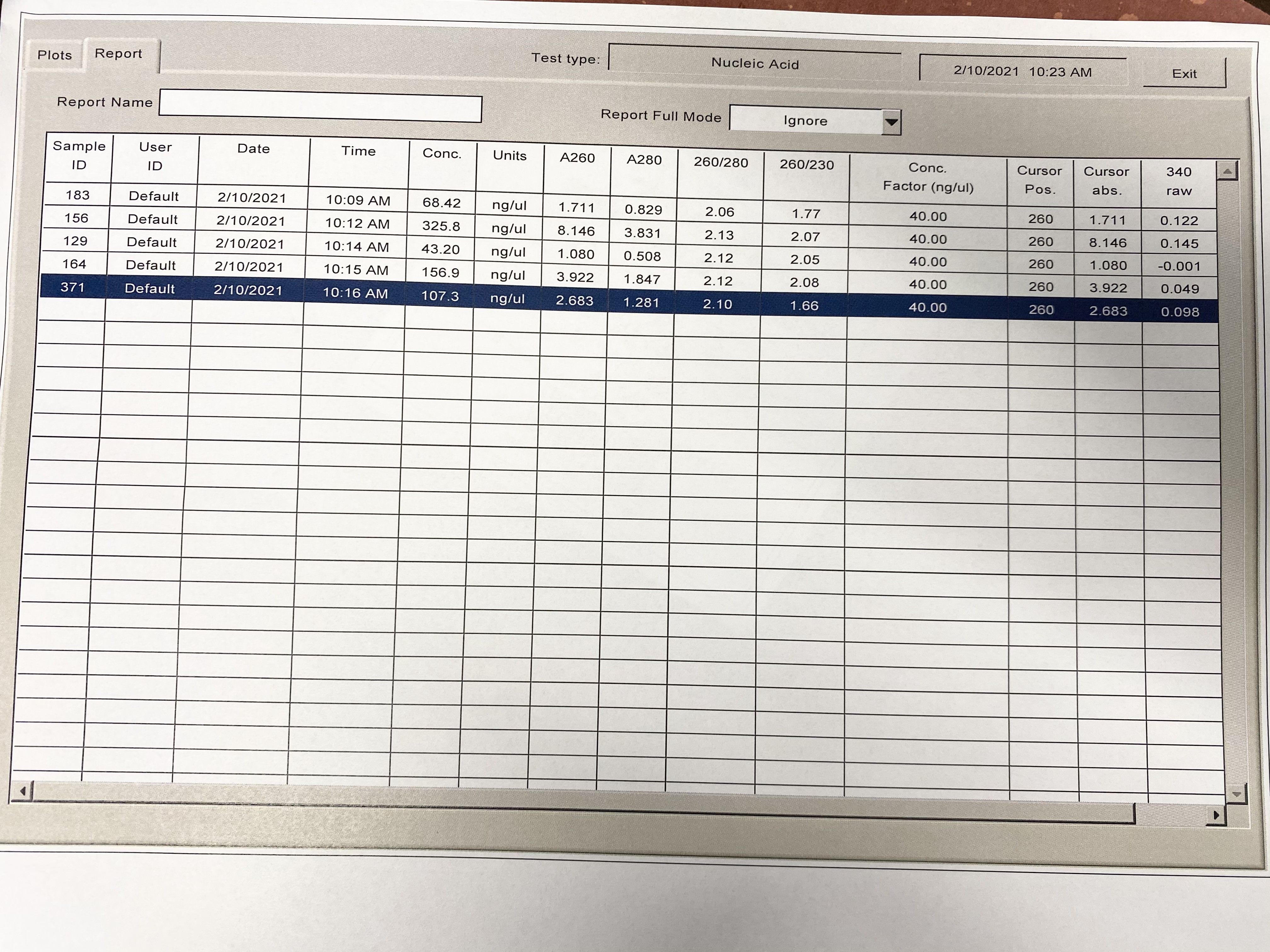
Even though 371 and 183 have a little low 260/230 values (should be ~2.00), the traces look pretty good, and the other samples have good ratios. They are from the same timepoint, so maybe that has something to do with it. It looks like there might not be a contamination issue with larvae samples.
I do still think I should check some RNA after doing a few test samples. Zymo no longer uses heated digestions (updated 1/27/2021), and suggests homogenization in something like a tissuelyser for any tissue samples. So I will probably have to do a slightly modified protocol than Erin’s.