Continuing Through-put Mo'orea Coral Extractions Two
Another 20 DNA only extractions from Porites and Pocillopora Corals from Mo’orea
Using the Zymo Quick-DNA Miniprep Plus kit
Sample Prep
| Sample # | Type |
|---|---|
| 42 | Pocillopora verrucosa |
| 46 | Pocillopora verrucosa |
| 60 | Pocillopora verrucosa |
| 161 | Pocillopora verrucosa |
| 165 | Pocillopora verrucosa |
| 169 | Pocillopora verrucosa |
| 182 | Pocillopora verrucosa |
| 183 | Pocillopora verrucosa |
| 188 | Pocillopora verrucosa |
| 202 | Massive Porites |
| 204 | Massive Porites |
| 214 | Massive Porites |
| 218 | Massive Porites |
| 226 | Massive Porites |
| 236 | Pocillopora verrucosa |
| 243 | Pocillopora verrucosa |
| 270 | Massive Porites |
| 307 | Massive Porites |
| 310 | Massive Porites |
| 311 | Massive Porites |
- New Beads were poured into sample tubes. The new beads should be easier to pipette the liquid out of them as the do not get sucked up by the p20
- Samples were homogenized by vortexing for ~60 seconds for all samples
- Most of the liquid from the tubes was removed by pipetting. I guessed that this was about 420µl. The tubes contained a small amount of liquid and un-homogenized tissue left, so 200µl of DNA/RNA shield was added to each tube and those were put back into the -20
- Following recommendations for samples in DNA/RNA Shield from the kit protocol, 210µl of Solid Tissue Buffer and 14µl of Proteinase K were added to each sample
- Samples were votexed, spun down, and incubated at 55 degrees C for 5 hours shaking at 600rpm
Following DNA extraction went along exactly as previous post
6µl of sample were aliquoted out for Qubit and Gel the next day to avoid thawing the stock.
Qubit
- Broad Range dsDNA Qubit protocol
- The Qubit was performed by Marygrace Trusdell
- All samples were read twice
| Sample | DNA Standard 1 (RFU) | DNA Standard 2 (RFU) | DNA 1 (ng/µl) | DNA 2 (ng/µl) | Average DNA |
|---|---|---|---|---|---|
| 42 | 196 | 16886 | 36.8 | 37.2 | 37 |
| 46 | 196 | 16886 | 52.8 | 51.8 | 51.3 |
| 60 | 196 | 16886 | 65.2 | 67.6 | 66.4 |
| 161 | 196 | 16886 | 52.8 | 54.2 | 53.5 |
| 165 | 196 | 16886 | 62.4 | 61.4 | 61.9 |
| 169 | 196 | 16886 | 40.2 | 38.6 | 39.4 |
| 182 | 196 | 16886 | 79.2 | 78.2 | 78.7 |
| 183 | 196 | 16886 | 47.6 | 48.8 | 48.2 |
| 188 | 196 | 16886 | 111 | 110 | 110.5 |
| 202 | 196 | 16886 | 15.2 | 14.8 | 15 |
| 204 | 196 | 16886 | 43.6 | 44 | 43.8 |
| 214 | 196 | 16886 | 48.4 | 46.8 | 47.7 |
| 218 | 196 | 16886 | 29.6 | 28.8 | 29.3 |
| 226 | 196 | 16886 | 18.3 | 17.8 | 18 |
| 236 | 196 | 16886 | 45.4 | 43.6 | 44.5 |
| 243 | 196 | 16886 | 77.2 | 76.4 | 76.7 |
| 270 | 196 | 16886 | 18.7 | 18.6 | 18.6 |
| 307 | 196 | 16886 | 13.7 | 12.8 | 13.4 |
| 310 | 196 | 16886 | 23 | 22.6 | 22.8 |
| 311 | 196 | 16886 | 17.4 | 17.4 | 17.4 |
67 and 220 were re-dos, so there might not have been that much left in the first place. I need to do a gel to see how the quality is.
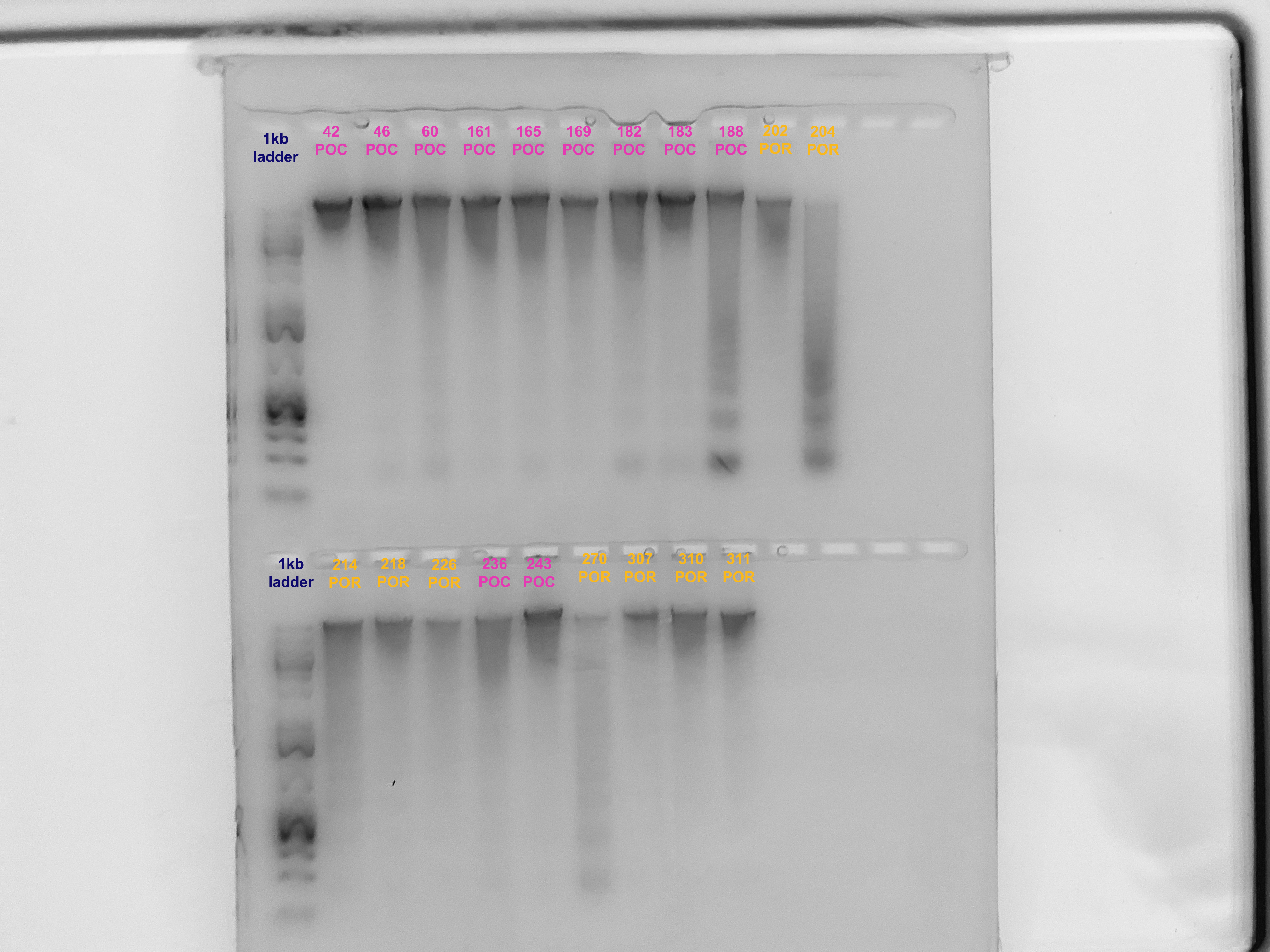
Gel Verification
- A 1.5% agarose gel was ran to check the integrity of the genomic DNA
- Following the PPP Lab protocol
- Samples 67 and 220 were not run on the gel
Written on April 22, 2019