Starting Through-put Mo'orea Coral Extractions
18 DNA only extractions from Porites and Pocillopora Corals from Mo’orea
Using the Zymo Quick-DNA Miniprep Plus kit
Sample Prep
| Sample # | Type |
|---|---|
| 5 | Massive Porites |
| 8 | Massive Porites |
| 49 | Pocillopora verrucosa |
| 50 | Pocillopora verrucosa |
| 67 | Massive Porites |
| 70 | Massive Porites |
| 192 | Pocillopora verrucosa |
| 198 | Massive Porites |
| 220 | Pocillopora verrucosa |
| 246 | Pocillopora verrucosa |
| 338 | Massive Porites |
| 364 | Pocillopora verrucosa |
| 172 | Pocillopora verrucosa |
| 176 | Pocillopora verrucosa |
| 186 | Pocillopora verrucosa |
| 384 | Pocillopora verrucosa |
| 405 | Massive Porites |
| 424 | Massive Porites |
- New Beads were poured into sample tubes. The new beads should be easier to pipette the liquid out of them as the do not get sucked up by the p20
- Samples were homogenized by vortexing for ~30 seconds for all samples
- Most of the liquid from the tubes was removed by pipetting. This was about 300µl. The tubes contained a small amount of liquid and un-homogenized tissue left, so 200µl of DNA/RNA shield was added to each tube and those were put back into the -20
- Following recommendations for samples in DNA/RNA Shield from the kit protocol, 150µl of Solid Tissue Buffer and 10µl of Proteinase K were added to each sample
- Samples were votexed, spun down, and incubated at 55 degrees C for 5 hours shaking at 600rpm
DNA Extraction
- Centrifuged all tubes at 12,000 rcf for 1 minute to pellet any debris and beads
- Removed supernatant into new 1.5mL tubes
- Added 1 volume (420µl) Genomic Binding Buffer to each tube, vortexed and spun down
- 700µl of sample was added to the kit spin column and centrifuged at 12,000 rcf for 1 minute
- Collection tubes were discarded
- The rest of the coral samples were run through the column in the same way
- Added 400µl DNA Pre-Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the flow through
- Added 700µl G-DNA Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the flow through
- Added 200µl G-DNA Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the collection tube
- Columns were transferred to 1.5mL tubes
- Added 50µl warmed 70 degrees C 10mM Tris-HCl directly to the column filter and incubated at room temp for 5 minutes
- Centrifuged for 1 minute at 12,000 rcf
- Repeated steps 11 and 12 one more time
- Stored tubes in fridge to qubit the next immediately
Qubit
- Broad Range dsDNA Qubit protocol
- Qubit was performed by Marygrace Trusdell
- All samples were read twice
| Sample | DNA Standard 1 (RFU) | DNA Standard 2 (RFU) | DNA 1 (ng/µl) | DNA 2 (ng/µl) | Average DNA |
|---|---|---|---|---|---|
| 5 | 232 | 20534 | 36 | 36.2 | 36.1 |
| 8 | 232 | 20534 | 24.6 | 24.2 | 24.4 |
| 49 | 232 | 20534 | 70 | 70.6 | 70.3 |
| 50 | 232 | 20534 | 73.4 | 73 | 73.2 |
| 67 | 232 | 20534 | 7.06 | 6.90 | 6.97 |
| 70 | 232 | 20534 | 38.8 | 39 | 38.9 |
| 192 | 232 | 20534 | 67.2 | 65.8 | 66.5 |
| 198 | 232 | 20534 | 40.8 | 40.8 | 40.8 |
| 220 | 232 | 20534 | 20.2 | 19.7 | 20 |
| 246 | 232 | 20534 | 36.2 | 34 | 35.1 |
| 338 | 232 | 20534 | 22.6 | 22 | 22.3 |
| 364 | 232 | 20534 | 46 | 50 | 48 |
| 172 | 232 | 20534 | 128 | 126 | 125 |
| 176 | 232 | 20534 | 86 | 86 | 86 |
| 186 | 232 | 20534 | 31.2 | 32.4 | 31.7 |
| 384 | 232 | 20534 | 54 | 54 | 54 |
| 405 | 232 | 20534 | 23.8 | 24.4 | 32.1 |
| 424 | 232 | 20534 | 48.6 | 47.8 | 47.2 |
Gel Verification
- A 1.5% agarose gel was ran to check the integrity of the genomic DNA
- Following the PPP Lab protocol
- Gel was run by Marygrace Trusdell
- Two samples are from Kevin Wong’s Astrangia homogenates
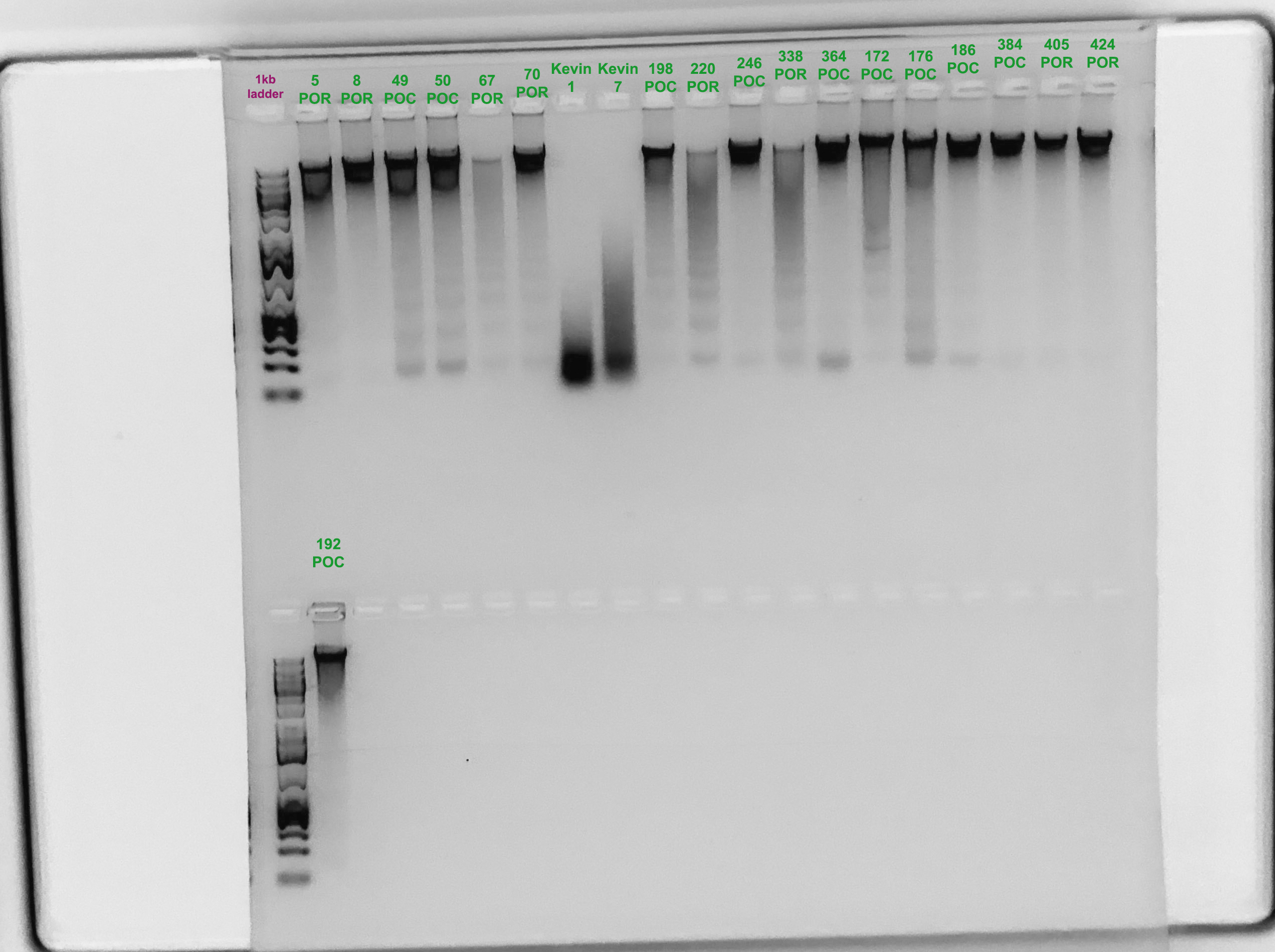
Samples 67, 220, and 338 don’t look like they are very good quality. I am going to try to re-extract from 67 and 220 because those are quite low. I should also try 338. This gel was run on the 18th, which is after I started the next extraction.
Written on April 15, 2019