Testing Quick-DNA Kit on Mo'orea Corals and Tide Pool Mytilus
I was not able to get good RNA at this time from the Zymo DNa/RNA kit. We are moving forward with trying a DNA only kit on the samples to save $, time, and reagents. This was also tried with two Mytilus californianus from the tide pool project that did not have good RNA with the other kit either.
Using this kit with some protocol modifications to fit our samples. All samples were prepped with separating beads from the liquid by the sieve because it gave the best DNA quantity last time.
Sample Prep: steps before both processing followed the same protocol
Corals
| Sample # | Site # | Date Collected | Type |
|---|---|---|---|
| 157 | 4 | 2018/03/13 | Pocillopora verrucosa |
| 157 | 4 | 2018/03/13 | Pocillopora verrucosa |
| 266 | 4 | 2018/03/13 | Massive Porites |
| 268 | 4 | 2018/03/13 | Massive Porites |
- Beads from bead tubes were poured into the sample tubes
- Samples were homogenized by vortexing for ~30 seconds. Porites samples were homogenized for an extra ~20 seconds
- Sieves/strainers were placed in new labeled 1.5mL tubes
- About 1/3 of the sample was poured into the sieves and centrifuged briefly as the lid does not fit on the centrifuge
- The liquid sample went through into the tube but most of the beads stayed in the sieve
- Repeated pouring the rest of the sample into the sieve and centrifuging quickly
- After all the sample possible went through the sieve, the sieves with beads and any un-homogenized tissue were labeled with the sample number, parafilmed on the top, and saved in the -20
- About 450µl was in each tube
- Following recommendations for samples in DNA/RNA Shield from the kit protocol, 225µl of Solid Tissue Buffer and 15µl of Proteinase K were added to each sample
- Samples were votexed, spun down, and incubated at 55 degrees C for 5 hours shaking at 600rpm
Mussels
| Sample | Location | Tide Pool | Date Collected | Extraction type |
|---|---|---|---|---|
| Mytilus 6 | Bob Creek | TP2 | 8/13/2017 | normal volume |
| Mytilus 7 | Bob Creek | TP2 | 8/13/2017 | double volume |
- Thawed mussels on ice. They were still icy during dissection
- New foil was put down for each mussel
- Forceps were cleaned with 10% bleach and DI water before use. A new scalpel blade was used for each mussel
- Mussels were opened with the scalpel and a small chunk of mantle was taken and put into a 1.5mL tube
- Mussels were given a number, put in a whirl pak, and returned to the -80
- The volumes in the kit protocol are very small (200µl) so for M7 I tried doubling those volumes to help get better recovery after bead homogenization
- Added 95µl of Nuclease free water and 95µl of Solid Tissue Buffer to M6
- Added 190µl of Nuclease free water and 190µl of Solid Tissue Buffer to M7
- Poured in beads and homogenized by vortexing for ~30 seconds
- Followed same steps as above with the sieve to collect the liquid
- Added 10µl Proteinase K to sample M6
- Added 20µl Proteinase K to sample M7
- Samples were votexed, spun down, and incubated at 55 degrees C for 5 hours shaking at 600rpm
DNA Extraction
- Centrifuged all tubes at 12,000 rcf for 1 minute to pellet any debris and beads that got through the sieve
- Removed supernatant into new 1.5mL tubes
- Added 1 volume Genomic Binding Buffer to each tube, vortexed and spun down
- Corals: 690µl
- M6: 200µl
- M7: 400µl
- 700µl, or all for the mussels, of sample was added to the kit spin column and centrifuged at 12,000 rcf for 1 minute
- Collection tubes were discarded
- The rest of the coral samples were run through the column in the same way
- Added 400µl DNA Pre-Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the flow through
- Added 700µl G-DNA Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the flow through
- Added 200µl G-DNA Wash Buffer, centrifuged at 12,000 rcf for 1 minute, and discarded the collection tube
- Columns were transferred to 1.5mL tubes
- Added 50µl warmed 70 degrees C 10mM Tris-HCl directly to the column filter and incubated at room temp for 5 minutes
- Centrifuged for 1 minute at 12,000 rcf
- Repeated steps 11 and 12 one more time
- Sample tubes were labeled and stored in the 4 degree fridge to quantify the next day
Qubit
- Broad Range dsDNA Qubit protocol
- DNA Qubit was preformed by Marygrace Trusdell
- All samples were read twice
| Sample | DNA Standard 1 (RFU) | DNA Standard 2 (RFU) | DNA 1 (ng/µl) | DNA 2 (ng/µl) | Average DNA |
|---|---|---|---|---|---|
| 157 | 203.63 | 22484 | 102 | 100 | 101 |
| 158 | 203.63 | 22484 | 178 | 178 | 178 |
| 266 | 203.63 | 22484 | 119 | 118 | 118.5 |
| 268 | 203.63 | 22484 | 84.2 | 83.4 | 83.6 |
| M6 | 203.63 | 22484 | 130 | 130 | 130 |
| M7 | 203.63 | 22484 | 128 | 127 | 127.5 |
These values are much higher than I have ever gotten out of the corals!!
Gel
- A 1.5% agarose gel was ran to check the integrity of the genomic DNA
- Following the PPP Lab protocol
- The first 6 samples are from this extraction, the last 4 samples are from the previous post
- Below the sample number is the amount of DNA in ng/µl from the Qubit
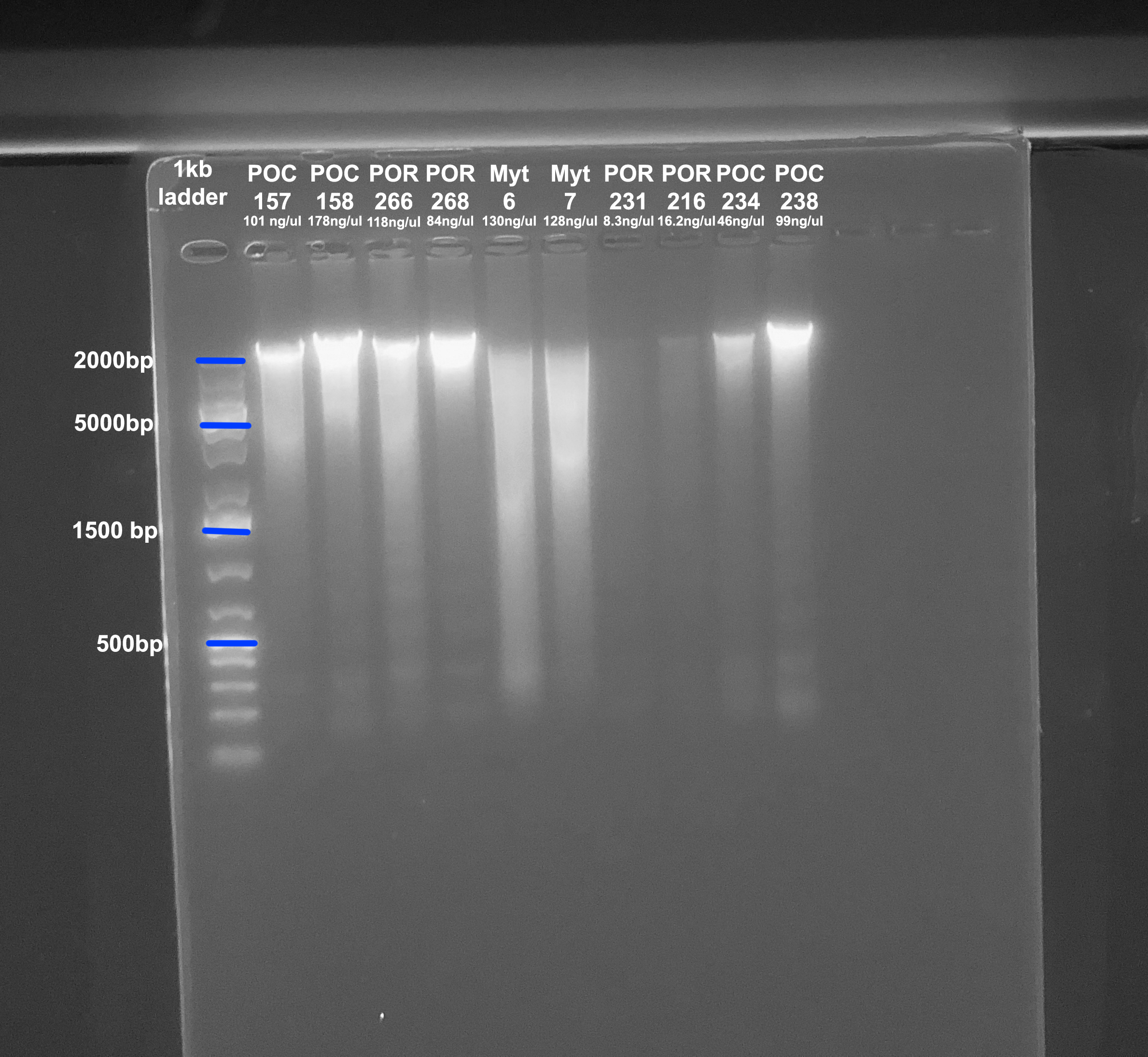
Mytilus DNA is not good quality here
Written on April 4, 2019