Bisulfite Conversion Test and TapeStation
Lightning Bisulfite Conversion of MBD Enriched Acropora Sperm Samples from Previous Post
Using the Zymo Lightning Methylation Kit, these same reagents are in the Zymo Pico Methyl Seq Kit
First step is to calculate how much DNA in each sample we have so that we can calculate the amount of non-methylated eColi DNA to spike into each sample to check for bisulfite conversion efficiency. We have that DNA diluted to 2.5ng/µl and we should spike 5ng for ever 100ng of sample.
| Sample | Total DNA |
|---|---|
| 221 NC | 253.7ng |
| 1431 NC | 735.3ng |
| 221 W | 5.76ng |
| 1431 W | 4.83ng |
| 221 C | 25.37ng |
| 1431 C | 15.69ng |
Wash (W) and not captured (NC) DNA were combined together. All tubes contained 21.5µl of sample, then the combined tubes held 43µl.
100ng of DNA of the not-captured samples was aliquoted out to make spike in calculations easy, and there is way more DNA than needed in those samples.
Preparation of samples to 20µl with spike in
221 Captured: Use whole 21.5µl and add 0.51µl of diluted eColi DNA
1431 Captured: Use whole 21.5µl and add 0.3µl of diluted eColi DNA *note here that 0.3 is probably not very accurate
221 Not-Captured Combined: 16.6µl of DNA, 3.4µl ultra pure water, and 2µl of eColi DNA
1431 Not-Captured Combined: 5.8µl of DNA, 14.2µl of ultra pure water, and 2µl of eColi DNA
Conversion Reaction
- 20µl of each tube was taken and pipetted into new PCR tubes, one for each sample
- 130µl of Lightning Conversion Reagent was added to each tube, vortexed, then spun down
- The 4 tubes were put into the thermocycler bisulfite conversion program (under mes user), which is 98 degrees C for 8 minutes, then 54 degrees C for 60 minutes, then 4 degree C hold
Conversion Cleanup
- Added 600µl of M-Binding buffer (lightning kit) to the provided Zymo-Spin IC Columns (4 of them, one per sample)
- Added the bisulfite treated DNA (150µl) to its respective column and inverted to mix
- Centrifuged at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 100µl M-wash buffer to each column and centrifuged at 12,000 rcf for 30 seconds
- Discarded flowthrough
- Added 200µl L-Desulfonation buffer to each column and let them sit at room temp for 15 minutes
- Began warming elution buffer to 56 degrees C
- Centrifuged for 30 seconds at 12,000 rcf and discarded flowthrough
- Added 200µl M-wash buffer, centrifuged for 30 seconds at 12,000 rcf and discarded flowthrough
- Repeated above wash step one time
- Transferred columns to 1.5mL tubes
- Added 8µl warmed elution buffer to the columns and let them sit for 1 minute
- Centrifuged for 30 seconds at 12,000 rcf
Took 1µl of captured samples for tapestation, used an RNA screentape because the DNA is now single stranded, but did not put the sample through the typical RNA denature program, just the buffer.
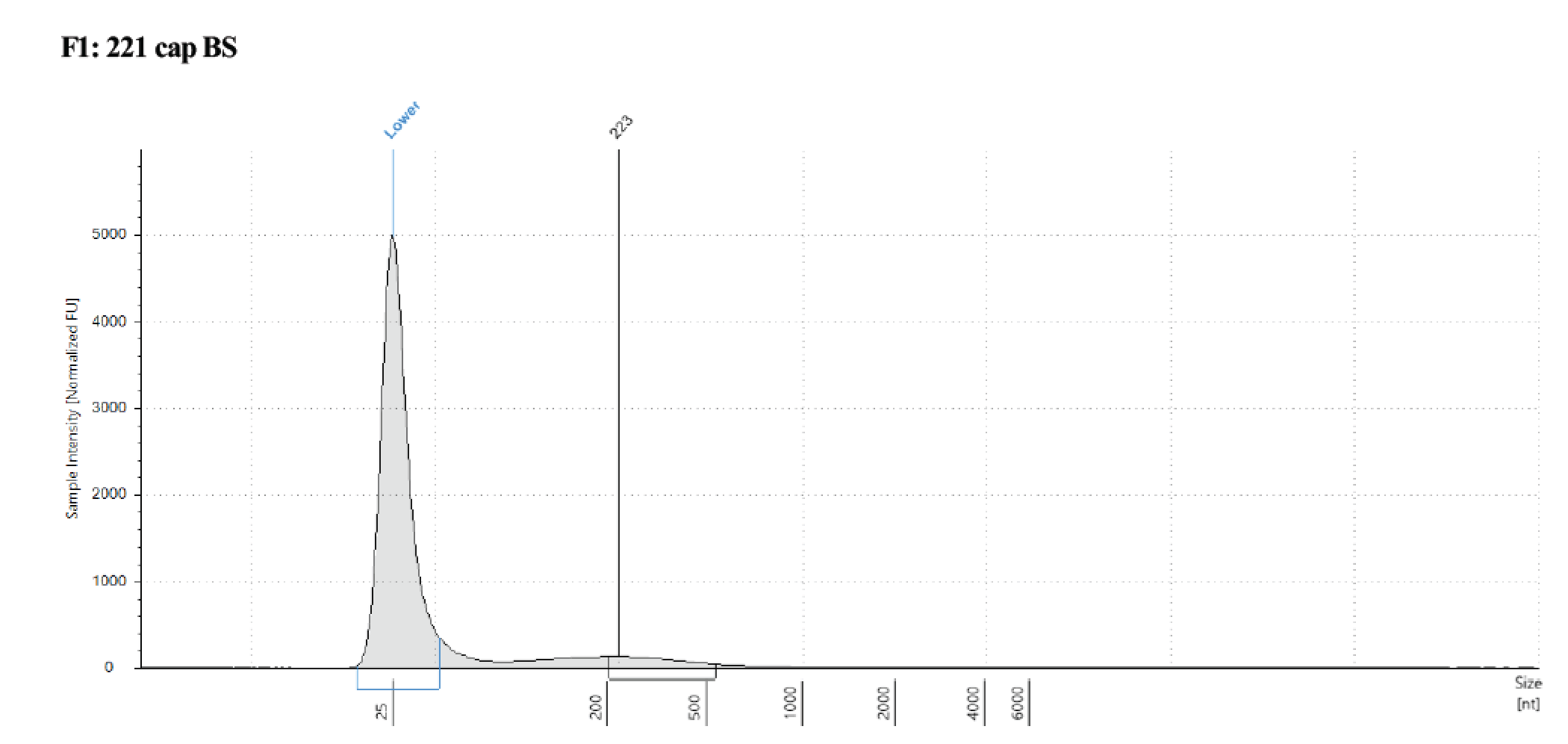
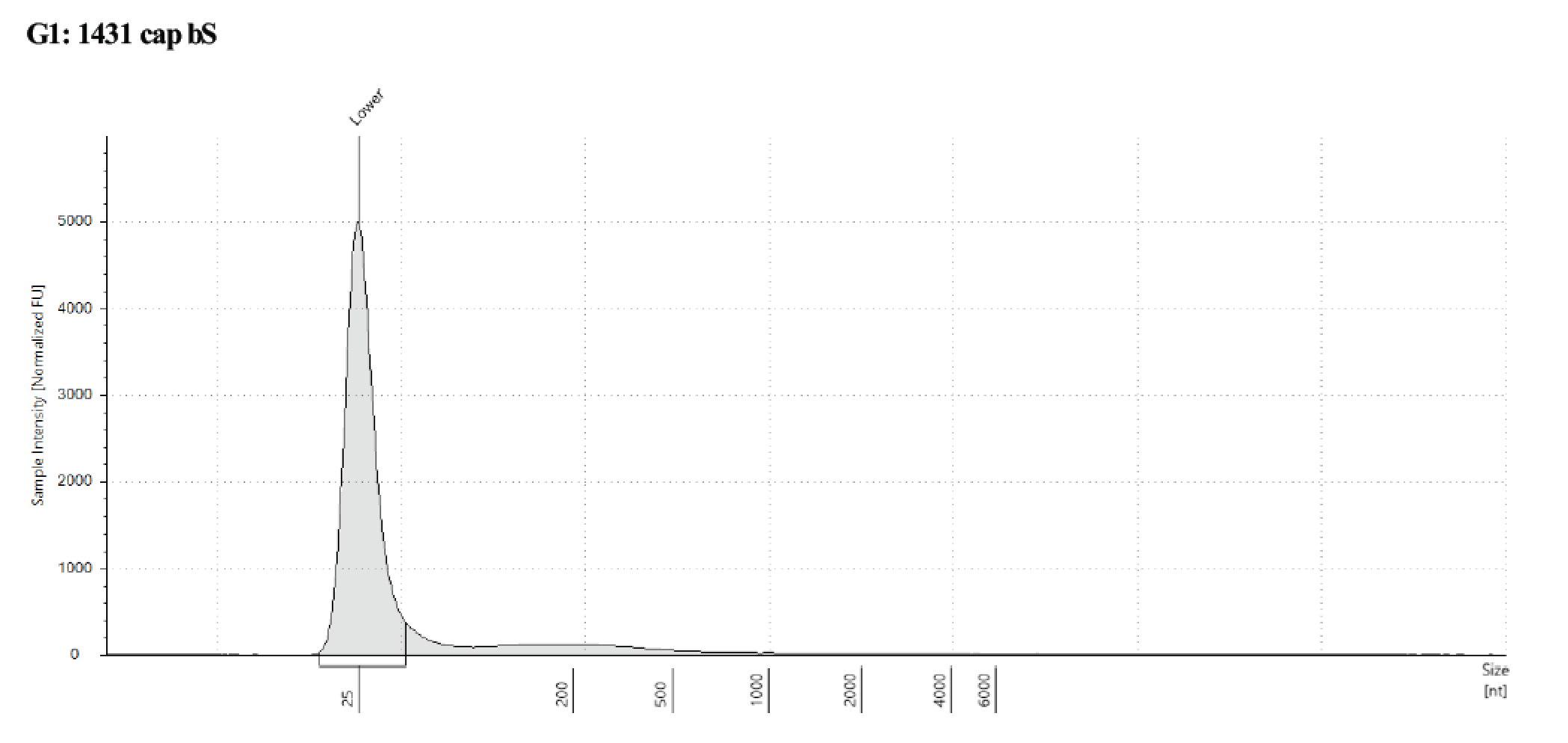
The size is hard to see, but around 200bp so that is good to go into the Pico MethylSeq kit for library prep
Full TS results include other samples of actual RNA