Montipora Larvae DNA/RNA Extraction 2
Using Larvae Extraction Protocol on 10 of Ariana’s Montipora Coral Larvae Stored in DNA/RNA Shield
Goal: Use optimized extraction method on 10 samples for Ariana’s project
Results: Good DNA and RNA quantity and quality, variable RNA purity
Take aways: Scaling up worked well, I am worried about the RNA purity in the last few samples. But the method was exactly the same for all of them, so I am not sure why it is bad in some samples. Maybe 3 will need to be extracted again because of the poor RNA quality, and maybe 12 will need to be extracted again to try to get DNA.
Sample Info:
| Sample | Timepoint | TP | Volume Shield in Storage | # Larvae |
|---|---|---|---|---|
| D3 | 1hpf | TP1 | 1mL | 250ul eggs |
| D10 | 38hpf | TP3 | 1mL note the spreadsheet says 500ul | 50ul |
| D12 | 65hpf | TP4 | 1mL note the spreadsheet says 500ul | 50ul |
| D16 | 65hpf | TP4 | 500ul | 50ul |
| D18 | 93hpf | TP5 | 500ul | 50ul |
| D20 | 93hpf | TP5 | 500ul | 50ul |
| D26 | 183hpf | TP7 | 300ul | 20-30 individuals |
| D28 | 183hpf | TP7 | 300ul | 20-30 individuals |
| D32 | 183hpf | TP9 | 300ul | 15-20 individuals |
| D39 | 231hpf | TP10 | 300ul | 15-20 individuals |
Based off of the sample info and how many larvae were present in each tube, save fractions were made for samples D3, D10, D12, D16, D18, and D20. No fractions were saved for D26, D28, D32, and D39.
| Sample | Volume Shield Added at Extraction | Save Fraction? | Saved Volume |
|---|---|---|---|
| D3 | 0 | yes | 500ul |
| D10 | 0 | yes | 500ul |
| D12 | 0 | yes | 500ul |
| D16 | 500ul | yes | 500ul |
| D18 | 500ul | yes | 500ul |
| D20 | 500ul | yes | 500ul |
| D26 | 200ul | no | - |
| D28 | 200ul | no | - |
| D32 | 200ul | no | - |
| D39 | 200ul | no | - |
Extraction
- Extraction protocol followed exactly
- Extraction proceeded with 500ul of each sample
- 500ul of DNA/RNA lysis buffer was used for each sample
- 1mL of 100% ethanol was used for each sample
QC 20210318
Qubit
- Broad Range dsDNA and High Sensitivity RNA Qubit protocol
- All samples were read twice
- One RNA sample needed to be read with the broad range, and I read D10 again as well because it was at the upper limit of the HS kit
BR DNA:
| Sample | Standard 1 | Standard 2 | DNA 1 ng/ul | DNA 2 ng/ul | Average ng/ul |
|---|---|---|---|---|---|
| D3 | 173 | 19382 | too low | - | - |
| D10 | - | - | 3.46 | 3.6 | 3.5 |
| D12 | - | - | too low | - | - |
| D16 | - | - | 24.4 | 24.4 | 24.4 |
| D18 | - | - | 24.2 | 24.2 | 24.2 |
| D20 | - | - | 33.8 | 33.4 | 33.6 |
| D26 | - | - | 39.6 | 39.8 | 39.7 |
| D28 | - | - | 20.8 | 21.2 | 21 |
| D32 | - | - | 5.88 | 6.02 | 5.95 |
| D39 | - | - | 6.12 | 5.86 | 5.99 |
HS RNA:
| Sample | Standard 1 | Standard 2 | RNA 1 ng/ul | RNA 2 ng/ul | Average ng/ul |
|---|---|---|---|---|---|
| D3 | 46.49 | 785 | too high | - | - |
| D10 | - | - | 104 | 104 | 104 |
| D12 | - | - | 84 | 84.4 | 84.2 |
| D16 | - | - | 96.2 | 95.8 | 96 |
| D18 | - | - | 55.2 | 55 | 55.1 |
| D20 | - | - | 68.2 | 68 | 68.1 |
| D26 | - | - | 45.4 | 45.4 | 45.4 |
| D28 | - | - | 28.8 | 29.6 | 29.2 |
| D32 | - | - | 14.3 | 14.4 | 14.35 |
| D39 | - | - | 13.2 | 12.8 | 13 |
BR RNA:
| Sample | Standard 1 | Standard 2 | RNA 1 ng/ul | RNA 2 ng/ul | Average ng/ul |
|---|---|---|---|---|---|
| D3 | 387 | 8079 | 562 | 560 | 561 |
| D10 | - | - | 126 | 126 | 126 |
NanoDrop
- Followed NanoDrop RNA Protocol
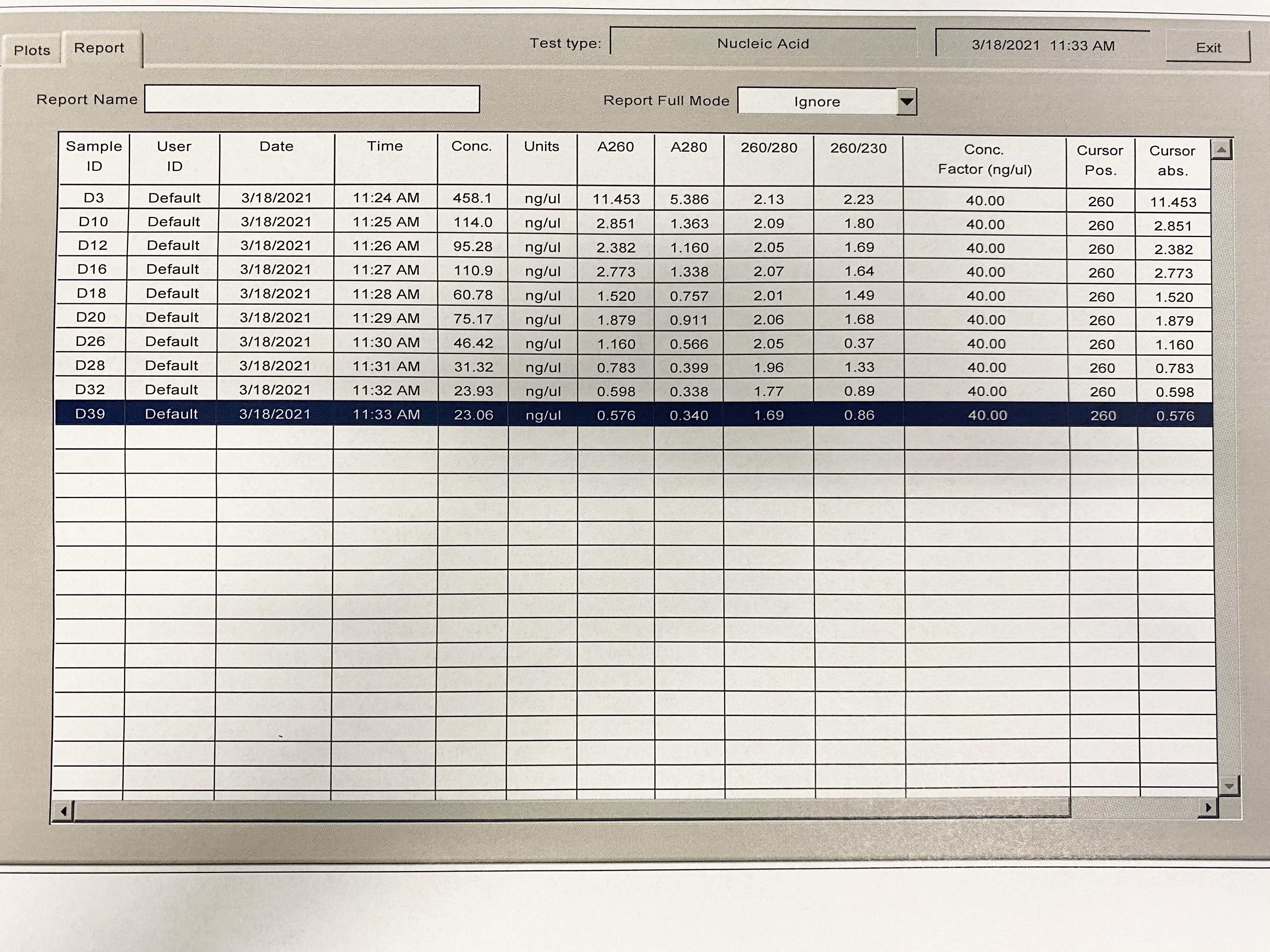
| Sample | 260/230 | 260/280 |
|---|---|---|
| D3 | 2.23 | 2.13 |
| D10 | 1.80 | 2.09 |
| D12 | 1.69 | 2.05 |
| D16 | 1.64 | 2.07 |
| D18 | 1.49 | 2.01 |
| D20 | 1.68 | 2.06 |
| D26 | 0.37 | 2.06 |
| D28 | 1.33 | 1.96 |
| D32 | 0.89 | 1.77 |
| D39 | 0.86 | 1.69 |
Full Results:
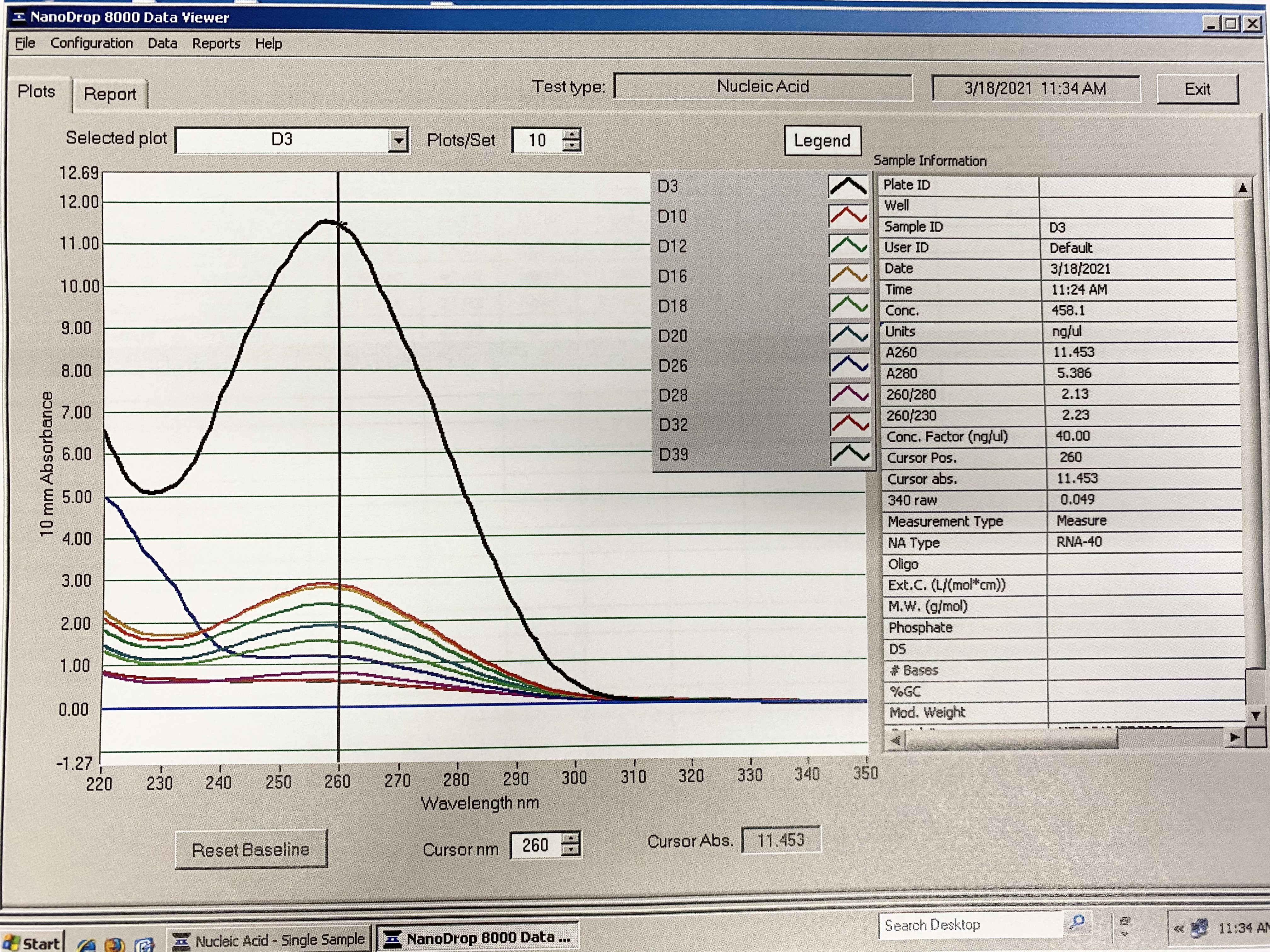
Traces:
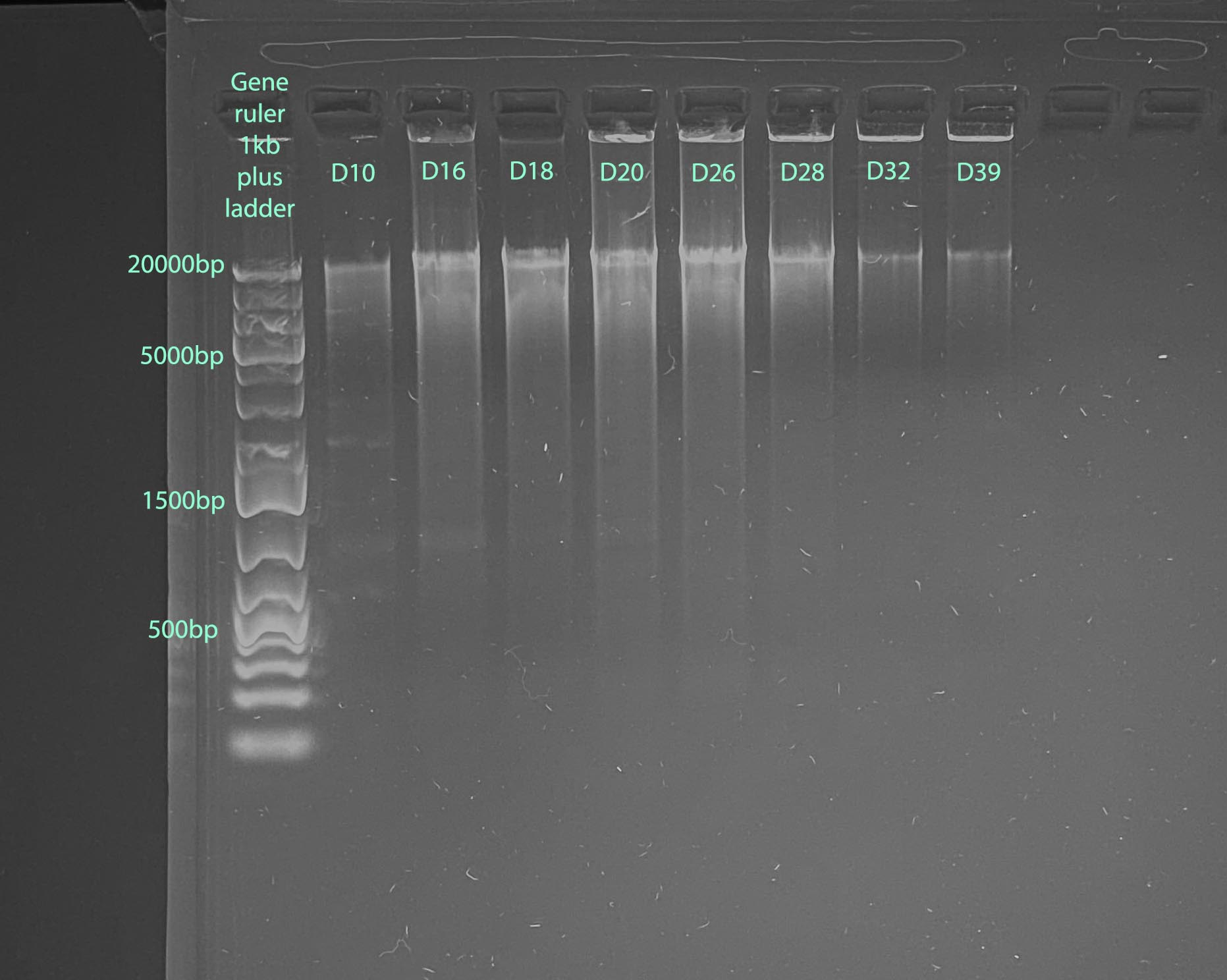
Gel
- 1% gel run for 1 hour at 100V
- Gel Protocol
TapeStation
- Followed RNA TapeStation protocol
| Sample | RIN score |
|---|---|
| D3 | 5.6 |
| D10 | 8.6 |
| D12 | 8.6 |
| D16 | 8.9 |
| D18 | 8.9 |
| D20 | 9.0 |
| D26 | 9.2 |
| D28 | 9.0 |
| D32 | 9.0 |
| D39 | 9.0 |