Montipora Larvae Test DNA/RNA Extraction 2021
Re-Testing Biomin Montipora Larvae/Eggs for DNA/RNA Extraction
Goal: Develop a good protocol for extracting high quality DNA and RNA and a good homogenization method for coral larvae/eggs
Results: No DNA for egg sample, pretty good DNA for the larvae sample. RNA quantity was very good, quality for the egg sample was very good, not as good for the larvae sample. 260/230 for egg sample not great.
Take-aways: This protocol might work, there was basically no issue with lipids in these two samples surprisingly. RNA is not great for the larvae, but similar to some of Erin’s extractions. However these samples were flash frozen and were not stored in DNA/RNA shield, which is now Ariana’s samples are stored.
Sample Info and Protocol Notes
- Samples are flash frozen eggs or larvae in 2mL screw cap tubes. Liquid volume in tubes is ~0
- Samples are Montipora and from the Biomineralization project
| Sample ID | Date | Timepoint | Treatment |
|---|---|---|---|
| 122 | 20180613 | egg | NA |
| 216 | 20180614 | 20:00 hours | xl |
- Followed homogenization method from adult coral DNA/RNA extraction protocol, however I used the full volume in the tubes to proceed in the extraction
Homogenization
- Took samples one at a time out of -80 and added 1mL of DNA/RNA shield
- Added 1/2 of the glass bead tubes to each sample tube
- Vortexed for 2 minutes on max speed
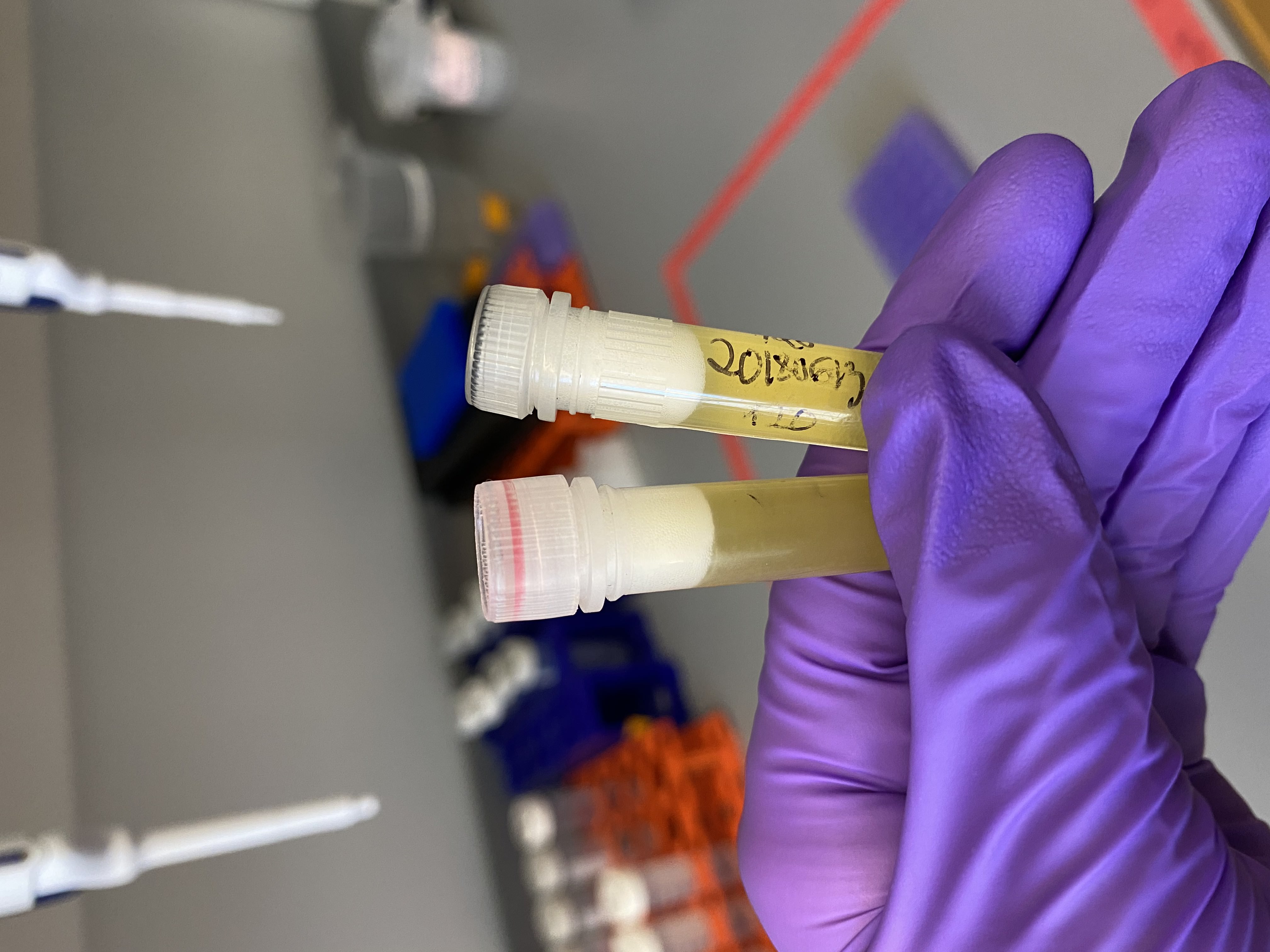
- Samples after vortexing, note they look completely homogenized and there seems to be no layer of lipids:
- Spun down tubes a few times to lower bubbles
- Added 100ul Pro K buffer to each tube
- Added 50ul Proteinase K to each tube
- Vortexed and spun down samples
- Transferred 1mL (most of the volume) to new 5mL tubes, avoiding beads. No debris or chunks were visible in the solution
DNA Extraction
- Warmed 10mM Tris HCl and ultrapure water in thermomixer to 70 degrees C
- Added equal volume of DNA/RNA lysis buffer to each 5mL tube (1mL)
- Vortexted and spun down tubes
- Sample tubes after buffer addition, still no lipid layer present:
122
216
- Added 700ul of that liquid to a yellow spin column for each sample
- Centrifuged at 16,000 rcf for 30 seconds
- Pipetted off the flowthrough into new 5mL tubes for each sample for RNA
- Repeated last three steps three more times until all the liquid had gone through the yellow spin columns for each sample
- Added 400ul prep buffer to each spin column
- Centrifuged at 16,000 rcf for 30 seconds
- Poured off flowthrough into kit waste beaker
- Added 700ul wash buffer to each spin column
- Centrifuged at 16,000 rcf for 30 seconds
- Poured off flowthrough into kit waste beaker
- Added 400ul wash buffer to each spin column
- Centrifuged for 2 minutes at 16,000 rcf
- Transferred the spin columns to new labeled 1.5mL tubes
- Added 50ul warmed 10mM Tris HCl to each column
- Incubated at room temp for 5 minutes
- Centrifuged at 16,000 rcf for 30 seconds
- Again added 50ul warmed 10mM Tris HCl to each column
- Incubated at room temp for 5 minutes
- Centrifuged at 16,000 rcf for 30 seconds
- Saved 10ul in strip tubes for QC, froze the rest in the -20
RNA Extraction
- Added equal volume fresh 100% ethanol to each of the flow through 5mL tubes from the DNA extraction (2mL)
- Vortexed and spun down tubes
- Added 700ul of that liquid to a green spin column for each sample
- Centrifuged at 16,000 rcf for 30 seconds
- Poured off flowthrough into kit waste beaker
- Repeated last three steps 5 times until all of the liquid had gone through the columns
- Added 400ul wash buffer to each spin column
- Centrifuged at 16,000 rcf for 30 seconds
- Poured off flowthrough into kit waste beaker
- Created the DNase mix:
- 2 * 75ul DNA digestion buffer = 150ul
- 2 * 5ul DNase I = 10ul
- Added 80ul of the DNase mix to each tube and incubated at room temp for 15 minutes
- Added 700ul wash buffer to each spin column
- Centrifuged at 16,000 rcf for 30 seconds
- Poured off flowthrough into kit waste beaker
- Added 400ul wash buffer to each spin column
- Centrifuged for 2 minutes at 16,000 rcf
- Transferred the spin columns to new labeled 1.5mL tubes
- Added 50ul warmed ultrapure water to each column
- Incubated at room temp for 5 minutes
- Centrifuged at 16,000 rcf for 30 seconds
- Again added 50ul warmed ultrapure water to each column
- Incubated at room temp for 5 minutes
- Centrifuged at 16,000 rcf for 30 seconds
- Saved 7ul in strip tubes for QC, the rest was frozen at -80 (Silver freezer, rack D column 3)
QC
Qubit
- Broad Range dsDNA and RNA Qubit protocol
- All samples were read twice
- DNA:
| Sample | Standard 1 | Standard 2 | DNA 1 ng/ul | DNA 2 ng/ul | Average ng/ul |
|---|---|---|---|---|---|
| 122 | 185 | 19673 | too low | - | - |
| 216 | - | - | 15.9 | 15.7 | 15.8 |
- RNA:
| Sample | Standard 1 | Standard 2 | RNA 1 ng/ul | RNA 2 ng/ul | Average ng/ul |
|---|---|---|---|---|---|
| 122 | 384 | 8304 | 98.8 | 98.2 | 96.5 |
| 216 | - | - | 242 | 240 | 241 |
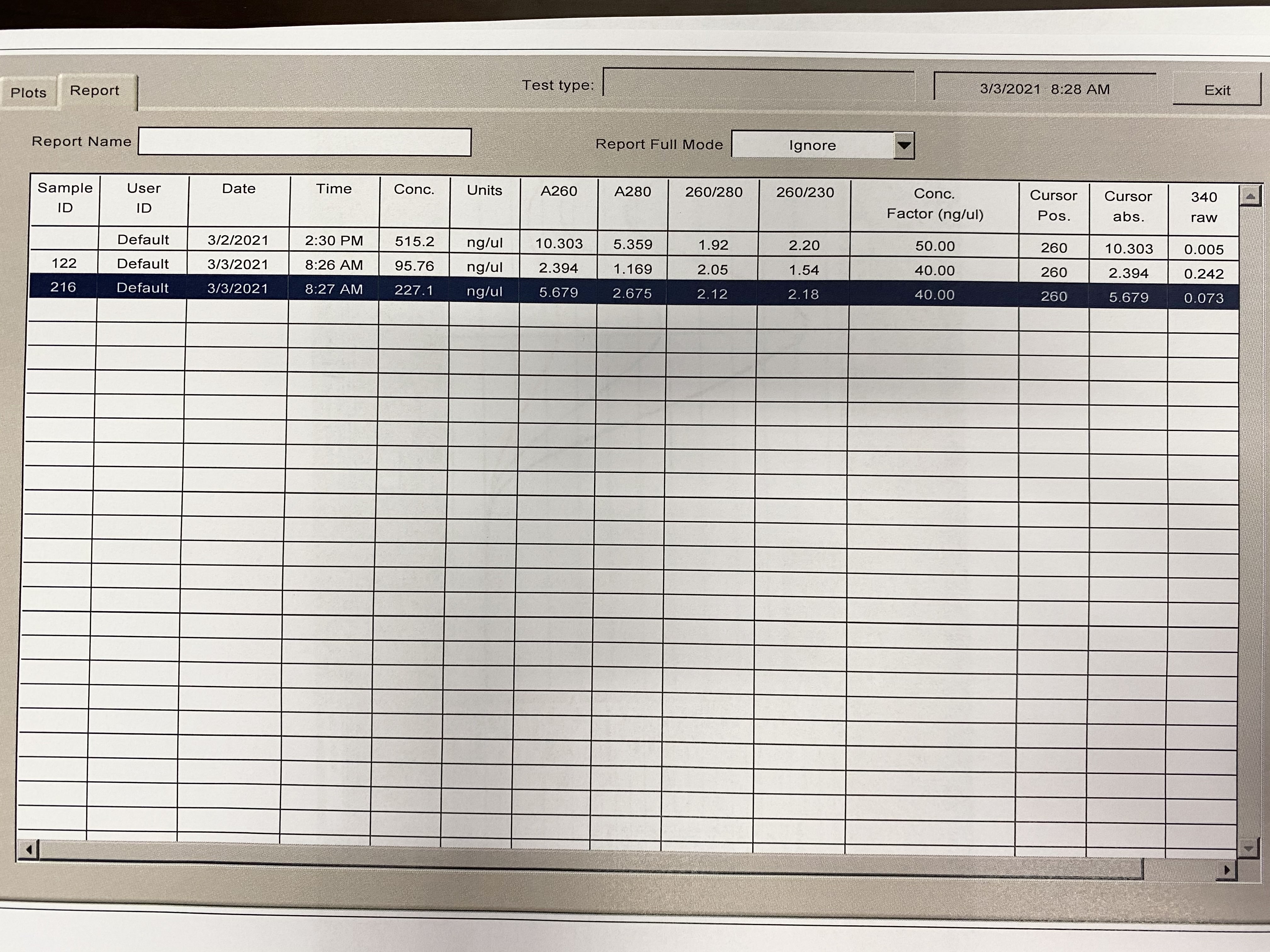
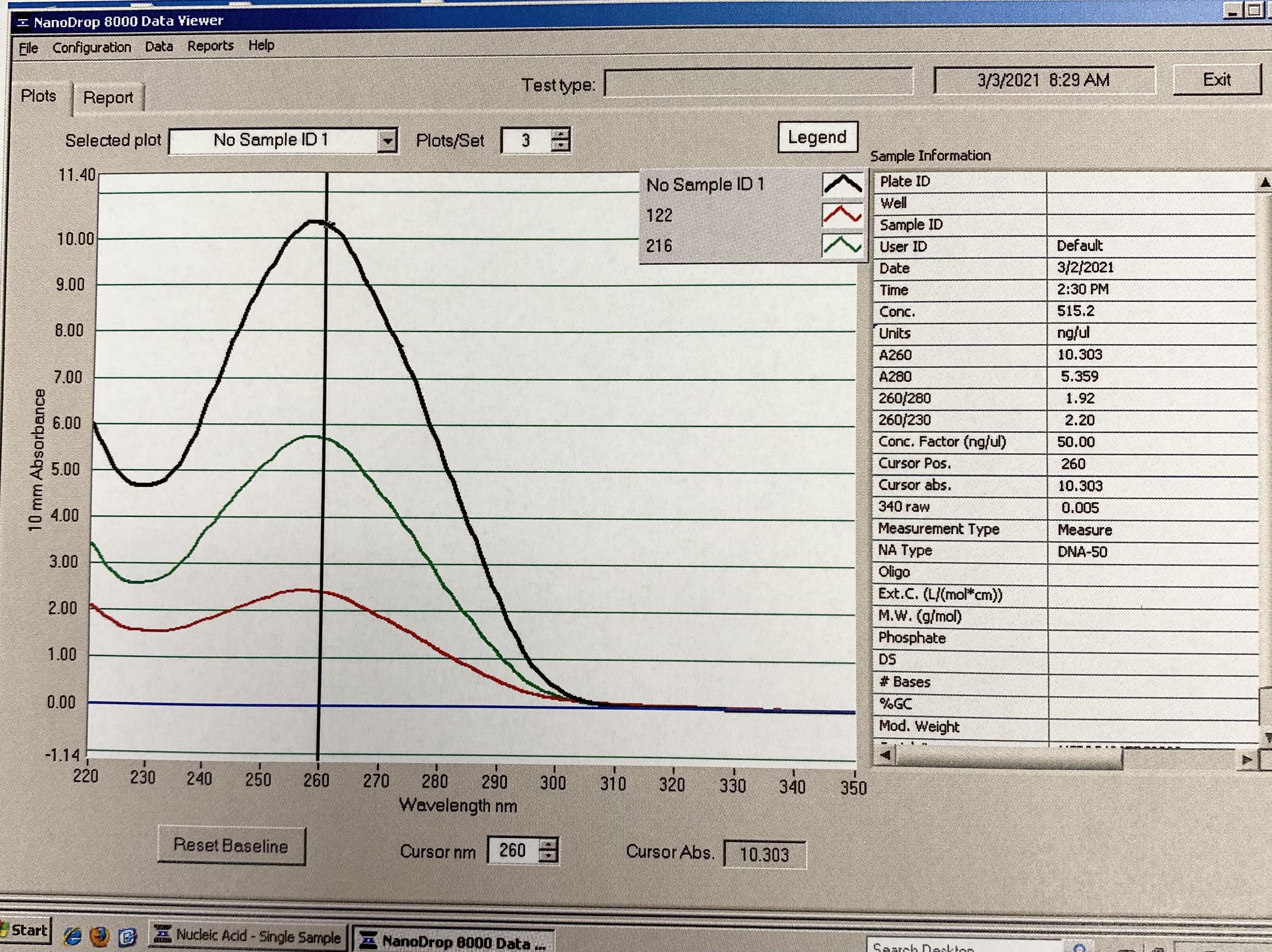
NanoDrop 20210304
- Followed NanoDrop RNA Protocol
- Note in images, for some reason the last sample someone had read before me came up in the table and plot, it doesn’t have a sample ID. Disregard those numbers
| Sample | 260/230 | 260/280 |
|---|---|---|
| 122 | 1.54 | 2.05 |
| 216 | 2.18 | 2.12 |
Full Results:
Traces:
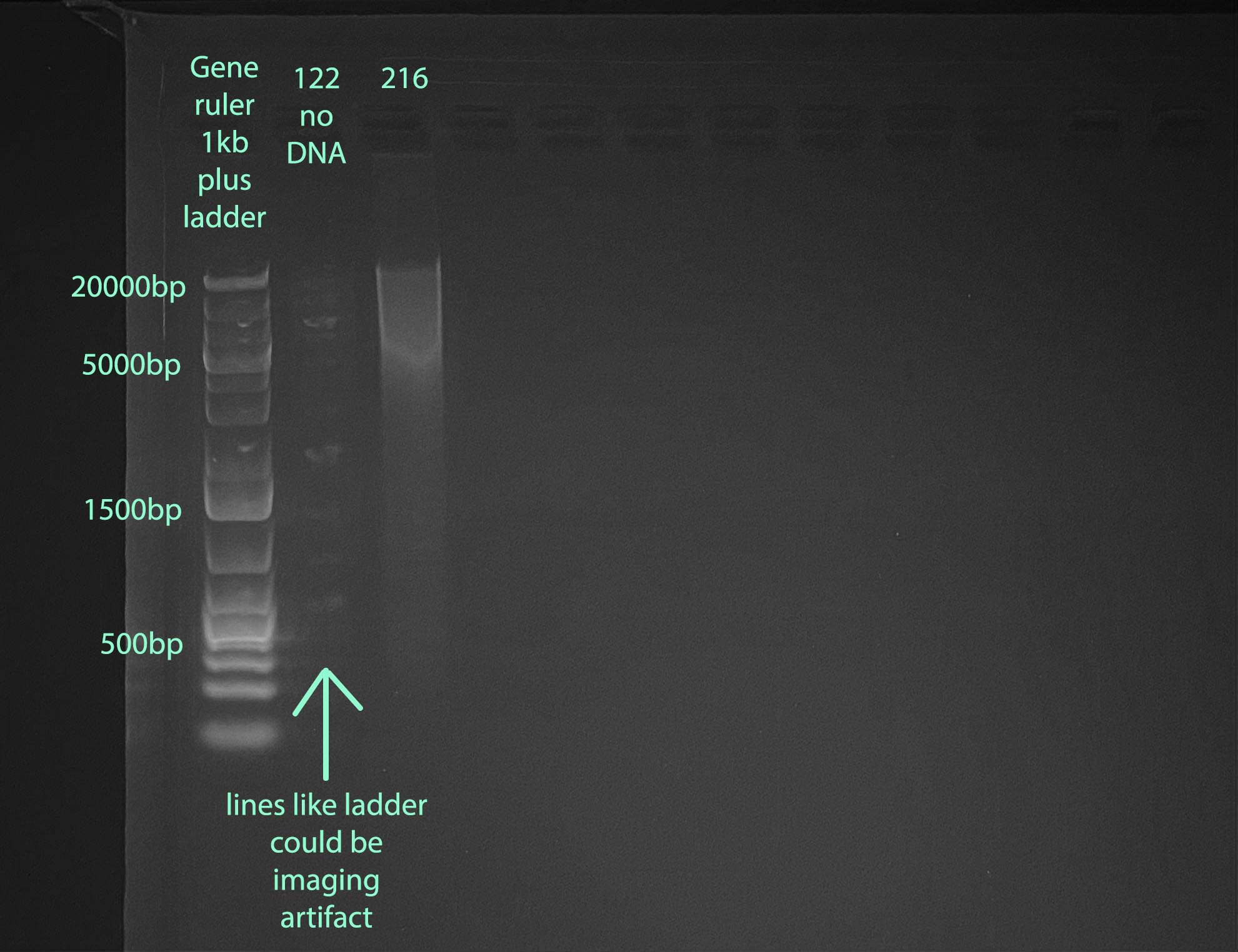
Gel
- 1% gel run for 1 hour at 100V
- Gel Protocol
- Note that I ran sample 122 before Qubiting so I did not know that there was no DNA. There is some weird fluorescence in that well, it could be imaging effects from the ladder, or a tiny amount of ladder got into that well
TapeStation
- Followed RNA TapeStation protocol
- Results link
Written on March 2, 2021