Montipora Larvae DNA/RNA Extraction 5
Using Larvae Extraction Protocol on 12 of Ariana’s Montipora Coral Larvae Stored in DNA/RNA Shield
Goal: Use optimized extraction method on 12 samples for Ariana’s project and elute the three samples above D29 in only 70ul of ultrapure water instead of 100ul
Results: Good DNA and RNA quantity and quality, RNA purity is not very good
Take aways: Purity of RNA is not very good, there seems to be no correlation with if the wash buffer is new or not, considering I used the same as the last extraction this time. Maybe I get worse purity the more samples I do at once
| Sample | Timepoint | TP | Volume Shield in Storage | # Larvae |
|---|---|---|---|---|
| D4 | 1hpf | TP1 | 1mL | 250ul eggs |
| D6 | 5hpf | TP2 | 1mL | 100ul |
| D7 | 5hpf | TP2 | 1mL | 100ul |
| D15 | 65hpf | TP4 | 500ul | 50ul |
| D21 | 163hpf | TP6 | 500ul | 50ul |
| D25 | 183hpf | TP7 | 300ul | 20-30 individuals |
| D27 | 183hpf | TP7 | 300ul | 20-30 individuals |
| D31 | 183hpf | TP9 | 300ul | 15-20 individuals |
| D33 | 231hpf | TP8 | 300ul | 20-30 individuals |
| D34 | 231hpf | TP8 | 300ul | 20-30 individuals |
| D35 | 231hpf | TP8 | 300ul | 20-30 individuals |
| D38 | 231hpf | TP10 | 300ul | 15-20 individuals |
Based off of the sample info and how many larvae were present in each tube, save fractions were made for samples D4, D6, D7, D15, and D21 only.
| Sample | Volume Shield Added at Extraction | Save Fraction? | Saved Volume |
|---|---|---|---|
| D4 | 0 | yes | 500ul |
| D6 | 0 | yes | 500ul |
| D7 | 0 | yes | 500ul |
| D15 | 500ul | yes | 500ul |
| D21 | 500ul | yes | 500ul |
| D25 | 200ul | no | - |
| D27 | 200ul | no | - |
| D31 | 200ul | no | - |
| D33 | 200ul | no | - |
| D34 | 200ul | no | - |
| D35 | 200ul | no | - |
| D38 | 200ul | no | - |
I eluted the RNA of D31, D33, D34, D35, and D38 in 70ul of ultrapure water instead of 100ul
Extraction
- Extraction protocol followed exactly
- Extraction proceeded with 500ul of each sample
- 500ul of DNA/RNA lysis buffer was used for each sample
- 1mL of 100% ethanol was used for each sample
- Samples D31, D33, D34, D35, and D38 were eluted with 40ul ultrapure water in the first elution and 30ul in the second
QC
Qubit
- Broad Range dsDNA and Borad Range RNA Qubit protocol
- All samples were read twice
BR DNA:
| Sample | Standard 1 | Standard 2 | DNA 1 ng/ul | DNA 2 ng/ul | Average ng/ul |
|---|---|---|---|---|---|
| D4 | 175 | 19024 | 2.02 | 2 | 2.01 |
| D6 | - | - | too low | ||
| D7 | - | - | too low | ||
| D15 | - | - | 26 | 26 | 26 |
| D21 | - | - | 43.4 | 43.4 | 43.4 |
| D25 | - | - | 28.8 | 28.6 | 28.7 |
| D27 | - | - | 26.4 | 26 | 26.2 |
| D31 | - | - | 8.02 | 7.9 | 8.01 |
| D33 | - | - | 21.6 | 21.4 | 21.5 |
| D34 | - | - | 24.2 | 24.2 | 24.2 |
| D35 | - | - | 25.8 | 25.6 | 25.7 |
| D38 | - | - | 12.2 | 12.3 | 12.25 |
BR RNA:
| Sample | Standard 1 | Standard 2 | RNA 1 ng/ul | RNA 2 ng/ul | Average ng/ul |
|---|---|---|---|---|---|
| D4 | 385 | 8145 | 278 | 276 | 277 |
| D6 | - | - | 140 | 140 | 140 |
| D7 | - | - | 179 | 177 | 178 |
| D15 | - | - | 66.6 | 66.2 | 66.4 |
| D21 | - | - | 53.6 | 53.6 | 53.6 |
| D25 | - | - | 28.8 | 28.6 | 28.7 |
| D27 | - | - | 27.8 | 27.8 | 27.8 |
| D31 | - | - | 20.2 | 19.6 | 19.9 |
| D33 | - | - | 32.6 | 32 | 32.3 |
| D34 | - | - | 32.2 | 32.4 | 32.3 |
| D35 | - | - | 30.6 | 31 | 30.8 |
| D38 | - | - | 18.2 | 18 | 18.1 |
NanoDrop
- Followed NanoDrop RNA Protocol
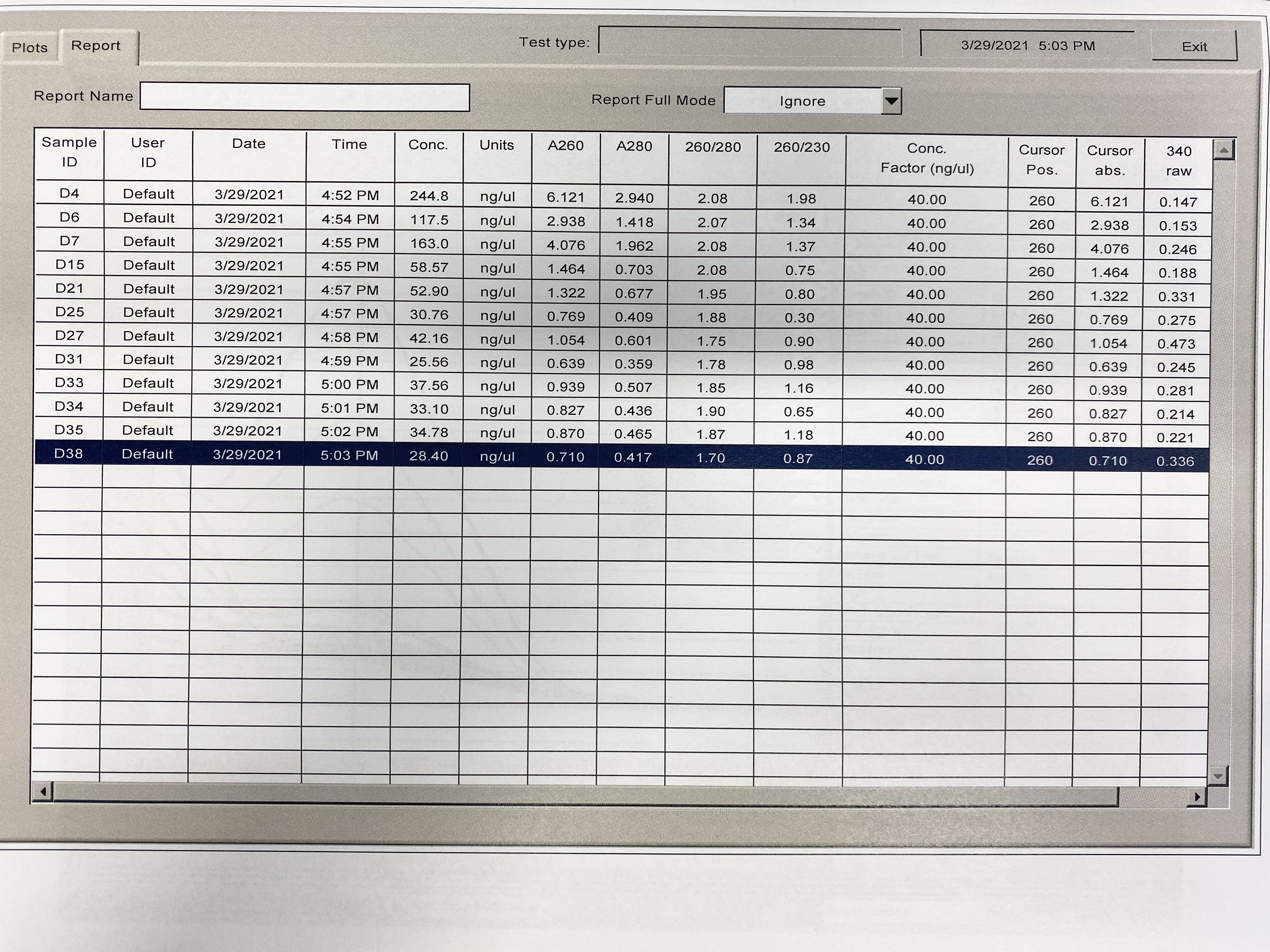
| Sample | 260/230 | 260/280 |
|---|---|---|
| D4 | 1.98 | 2.08 |
| D6 | 1.34 | 2.07 |
| D7 | 1.37 | 2.08 |
| D15 | 0.75 | 2.08 |
| D21 | 0.80 | 1.95 |
| D25 | 0.30 | 1.88 |
| D27 | 0.90 | 1.75 |
| D31 | 0.98 | 1.78 |
| D33 | 1.16 | 1.85 |
| D34 | 0.65 | 1.90 |
| D35 | 1.18 | 1.87 |
| D38 | 0.87 | 1.70 |
Full Results:
Traces:

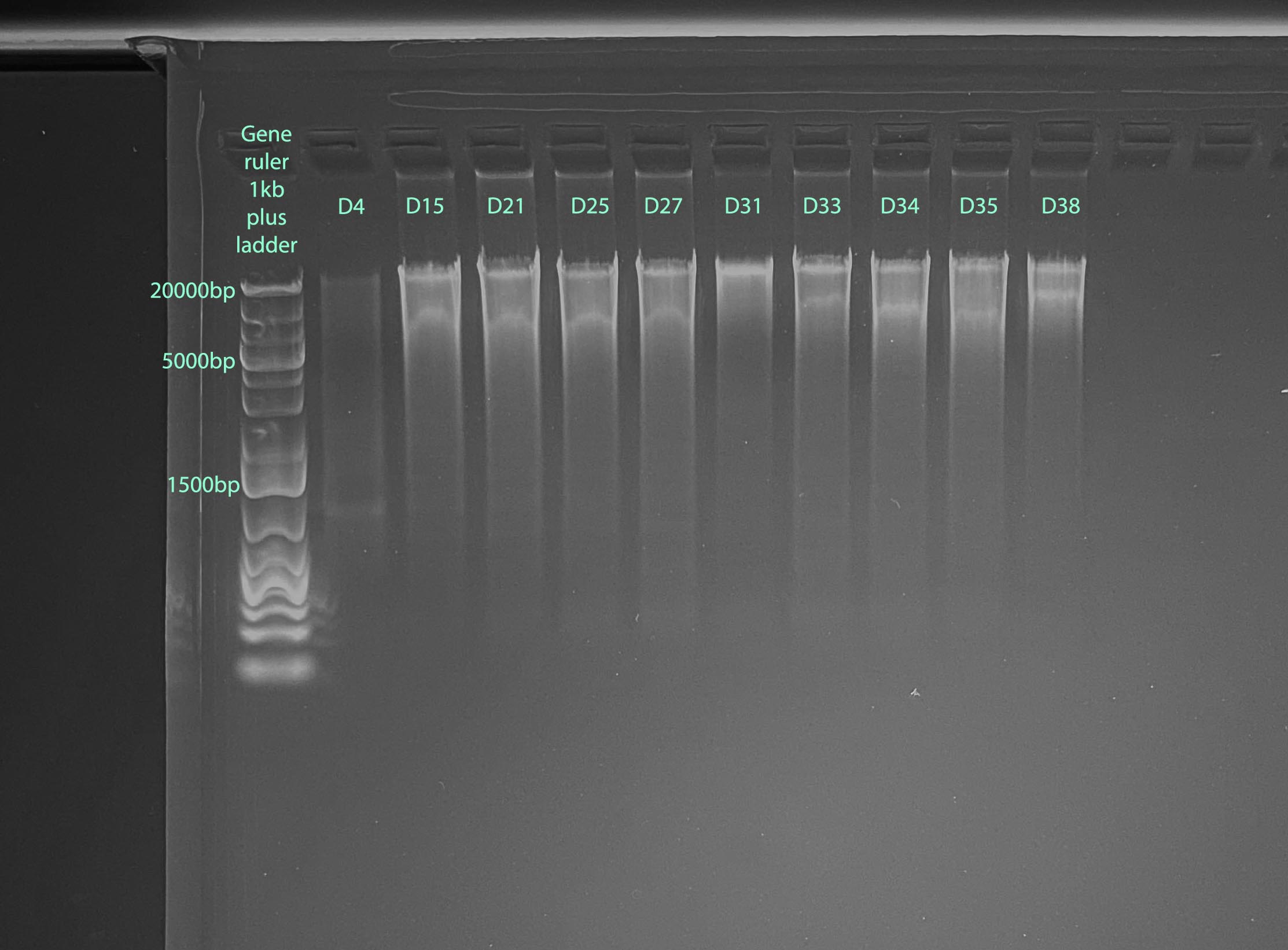
Gel
- 1% gel run for 1 hour at 100V
- Gel Protocol
TapeStation
- Followed RNA TapeStation protocol
- Used two tapes because one was open
| Sample | RIN score |
|---|---|
| D4 | 7.8 |
| D6 | 7.9 |
| D7 | 8.0 |
| D15 | 7.9 |
| D21 | 8.2 |
| D25 | 8.4 |
| D27 | 8.5 |
| D31 | 7.8 |
| D33 | 8.6 |
| D34 | 8.9 |
| D35 | 8.7 |
| D38 | NA/too low to read, looks good though |