Electroporating pSPIN-BAC Cells with DiNV DNA and Treating Resulting Cells With DNase to Remove Outside DNA
Our thought is that the DNA is getting stuck to the outside of the bacteria even with washing, so that we can’t tell if the DiNV DNA is actually getting inside the cells during the electroporation. But if we treat the bacteria with DNase, then DNA outside of the cells should be degraded, and virus DNA remaining inside of the cells should still be there.
Electroporation
- Prepared 4 1.5mL tubes with 975ul SOC buffer
- Using sample 26-exo
- Thawed 2 tubes of pSPIN-BAC electrocompetent cells on ice
- Brought up to Chandler lab:
- ice with bacteria and DNA
- pipettes and tips
- bacterial tip waste
- tape
- 20 1.5mL tubes
- tubes with SOC buffer
- Placed 4 SOC buffer tubes in the 30C incubator
- Placed 2 electroporation EC2 cuvettes on ice
- pSPIN-BAC + DNA no electro - tube 14A and 14B
- Added 25ul pSPIN-BAC cells to a tube on ice labeled 14A
- Added 2ul 26-exo DNA
- Immediately added 975ul of 37C SOC buffer
- Transferred 490ul to a 1.5mL tube labeled “14B”
- Placed 14B on ice
- Placed 14A in the 30C shaking incubator 1 hour
- pSPIN-BAC + DNA no electro - tube 15A and 15B (for DNase treatment)
- Added 25ul pSPIN-BAC cells to a tube on ice labeled 15A
- Added 2ul 26-exo DNA
- Immediately added 975ul of 37C SOC buffer
- Transferred 490ul to a 1.5mL tube labeled “15B”
- Placed 15B on ice
- Placed 15A in the 30C shaking incubator 1 hour
- pSPIN-BAC + DNA WITH electro - tube 16A and 16B
- Added 25ul pSPIN-BAC cells to a cuvette on ice
- Added 2ul 26-exo DNA
- Electroporated on EC-2 settings
- Immediately added 975ul of 30C SOC buffer
- Transferred 490ul to two 1.5mL tube labeled “16A and 16B”
- Placed 16B on ice
- Placed 16A in the 30C shaking incubator 1 hour
- pSPIN-BAC + DNA WITH electro - tube 17A and 17B (for DNase treatment)
- Added 25ul pSPIN-BAC cells to a cuvette on ice
- Added 2ul 26-exo DNA
- Electroporated on EC-2 settings
- Immediately added 975ul of 30C SOC buffer
- Transferred 490ul to two 1.5mL tube labeled “17A and 17B”
- Placed 17B on ice
- Placed 17A in the 30C shaking incubator 1 hour
B Samples
- All B samples were taken to 4055 where room temp LB was waiting
- Spun down tubes 3 min at 6,000g
- Discarded supernatant
- Resuspended pellets in 300ul LB
- Spun down tubes 3 min at 6,000g
- Discarded supernatant
- Tubes 14B and 16B were frozen at -80
- Tubes 15B and 17B were used for DNase treatment
B Sample DNase treatment
- Using promega DNase and reagents I found in the fridge
- Added 40ul of molecular grade water to each tube
- Added 5ul of 10X reaction buffer
- Added 2ul of DNase
- Flicked to mix then spun down
- Placed in the heat block for 30 minutes at 37C
- Added 5ul DNase stop solution and flicked to mix and spun down
- Incubated at 65C heat block for 10 minutes
- Centrifiged for 3 min 6000g
- Removed supernatant (saved)
- Froze pellets at -80C
A Samples
- All A samples were taken to 4055 where room temp LB was waiting after incubation for 1 hour
- Spun down tubes 3 min at 6,000g
- Discarded supernatant
- Resuspended pellets in 300ul LB
- Spun down tubes 3 min at 6,000g
- Discarded supernatant
- Tubes 14A and 16A were frozen at -80
- Tubes 15A and 17A were used for DNase treatment
A Sample DNase treatment
- Added 40ul of molecular grade water to each tube
- Added 5ul of 10X reaction buffer
- Added 2ul of DNase
- Flicked to mix then spun down
- Placed in the heat block for 30 minutes at 37C
- Added 5ul DNase stop solution and flicked to mix and spun down
- Incubated at 65C heat block for 10 minutes
- Centrifiged for 3 min 6000g
- Removed supernatant (saved)
- Froze pellets at -80C
DNA Extraction
- All samples were taken out of the -80 and put on ice
- DNA extraction was done exactly as described in a previous post
Qubit and Dilution
- I wanted to make sure all the samples were at the same concentration before the PCRs
- Qubit followed the protocol
- Then samples were diluted to 1ng/ul for PCR (note C and D samples were supernatants from DNase treatment and shouldn’t really have anything)
| tube | ng/ul | ul DNA for 1ng/ul | ul hydration solution |
|---|---|---|---|
| 14A | 38 | 2 | 74 |
| 15A | 10.2 | 2 | 18.4 |
| 16A | 5.86 | 2 | 9.72 |
| 17A | 3 | 3 | 6 |
| 14B | 4.47 | 3 | 11.18 |
| 15B | 1.85 | 4 | 4 |
| 16B | 4.14 | 3 | 9.42 |
| 17B | 0.825 | NA | NA |
| 15C | NA | NA | NA |
| 15D | NA | NA | NA |
| 17C | NA | NA | NA |
| 17D | NA | NA | NA |
PCRs on DNA Extracts
- The point was test all of the DNA samples for if they have retained DiNV DNA with PCR
- 4 PCRs were run on the samples, and the process followed the general PCR protocol completely. Master mix volumes are listed here:
| reagent | p47 | 16S | lef 9 | lef 4 |
|---|---|---|---|---|
| GoTaq | 75ul | 75ul | 75ul | 75ul |
| F primer | 3.75ul | 3.75ul | 3.75ul | 3.75ul |
| R primer | 3.75ul | 3.75ul | 3.75ul | 3.75ul |
| molecular grade water | 52.5ul | 52.5ul | 52.5ul | 52.5ul |
- All PCR programs were run for 30 cycles except for p47 which was then re-run for 30 cycles, and program information can be found here
- 1% gels were run at 90V for 45 minutes to resolve the bands:
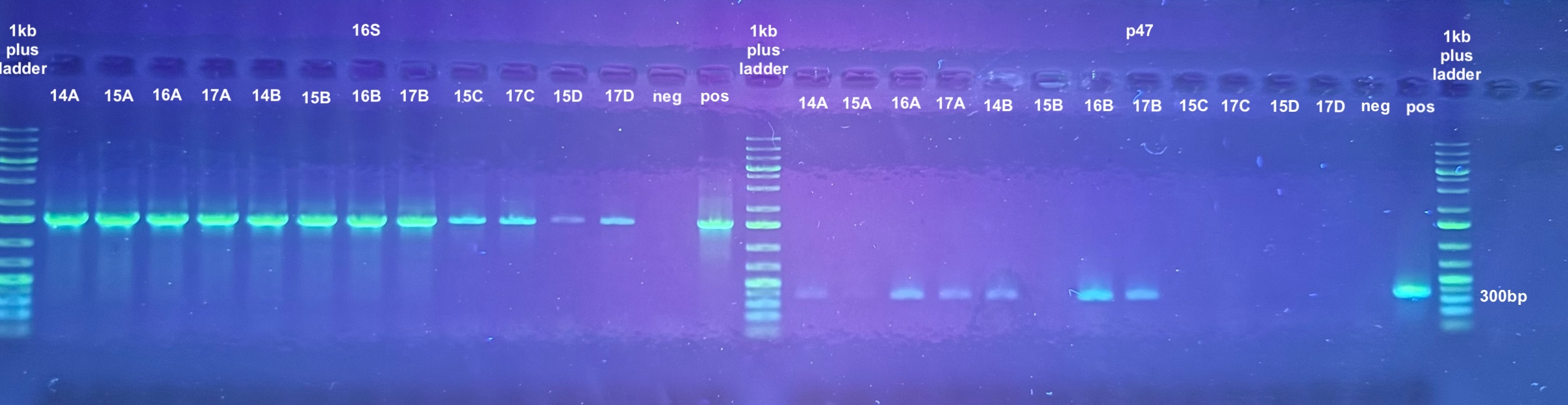
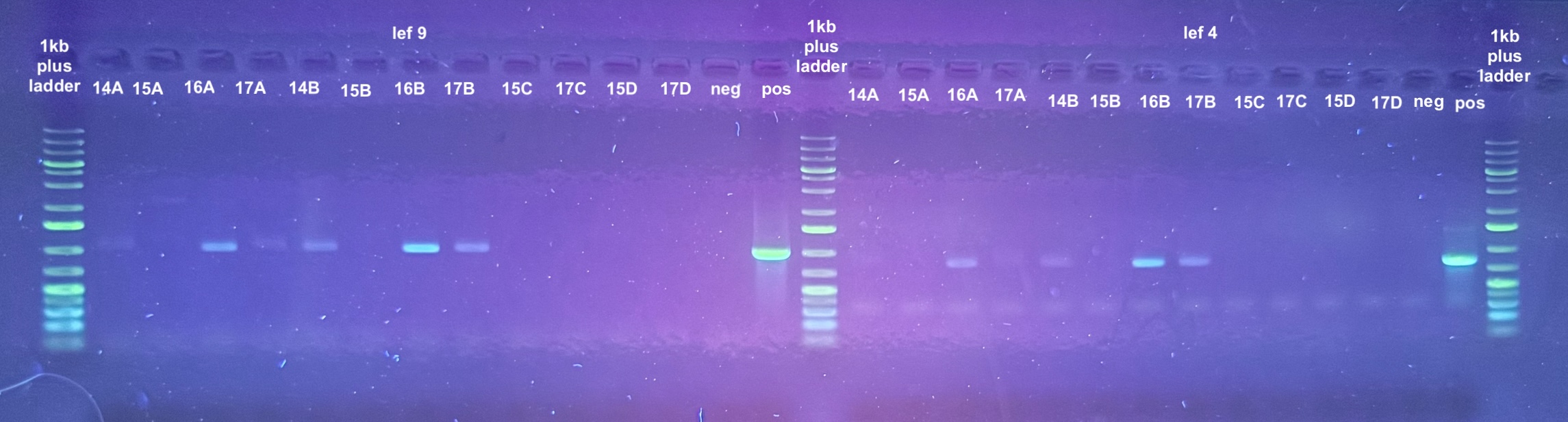
| sample number | electroporation | incubation | DNase treatment | p47 result | lef 9 result | lef 4 result |
|---|---|---|---|---|---|---|
| 14 A | no | yes | no | yes | maybe | no |
| 15 A | no | yes | yes | no | no | no |
| 16 A | yes | yes | no | yes | yes | yes |
| 17 A | yes | yes | yes | yes | faint band | faint band |
| 14 B | no | no | no | yes | yes | yes |
| 15 B | no | no | yes | no | no | no |
| 16 B | yes | no | no | yes | yes | yes |
| 17 B | yes | no | yes | yes | yes | yes |
| 15 C | no | yes | leftover solution from post-DNase spindown | No | No | No |
| 17 C | yes | yes | leftover solution from post-DNase spindown | No | No | No |
| 15 D | no | no | leftover solution from post-DNase spindown | No | No | No |
| 17 D | yes | no | leftover solution from post-DNase spindown | No | No | No |
This does indicate that DNA does get inside the cells (17A and 17B)