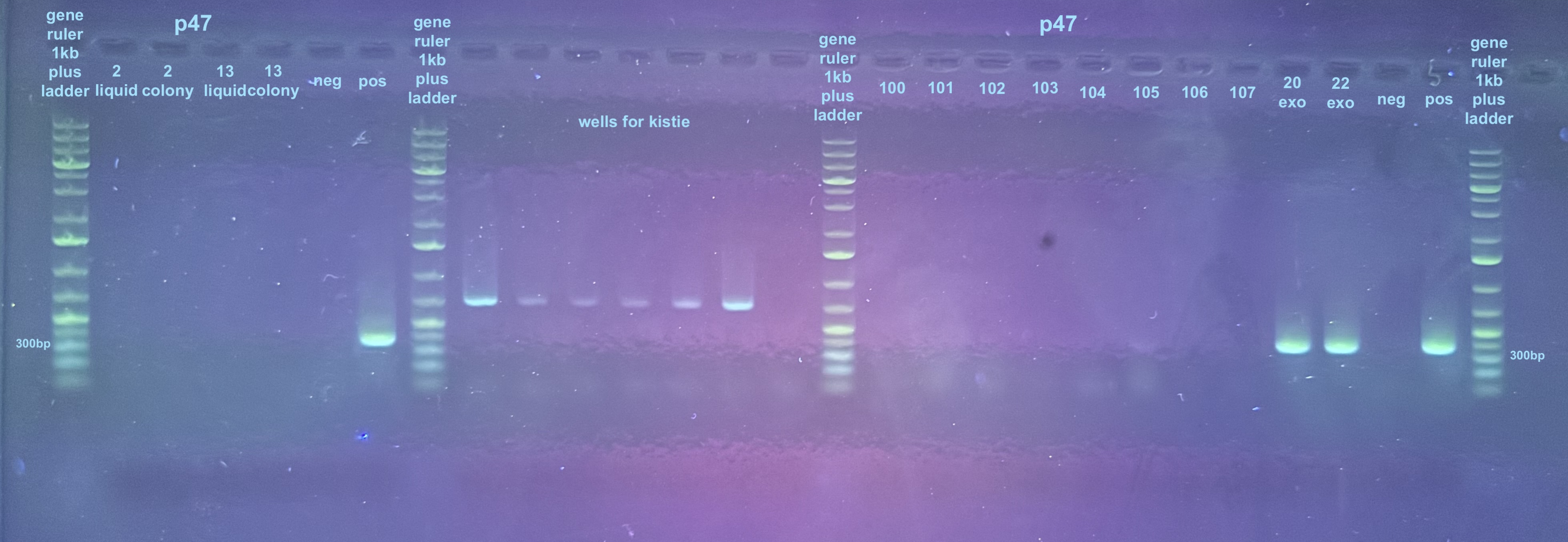
Growth, DNA Extraction, and p47 PCRs on Colonies 2 and 13 from 1st Electroporation
There were faint slight double bands on colonies 2 and 13 from the first electroporation attempt that looked like something amplifed for p47. Colony 2 is a control so that would not be good if it was virus. Colony 13 is one that got the virus DNA. I wanted to grow up the colonies in liquid culture and do DNA extractions and check that DNA for p47 amplification.
On 1/30 overnight liquid cultures of 2mL were made from colonies 2 and 13, one tube was the remaining liquid left from the colony swish in water, and the other took another pipette dab at the original colony plate (these colonies were marked on the plate). The 2mL cultures were first mixed with 0.8ul of stock chloramphenicol. These were grown overnight, and when there was not time the next day to do the DNA extraction, 10ul from this overnight culture was used to seed another overnight culture.
Samples:
| number | colony | type |
| 1 | 2 | liquid |
| 2 | 2 | colony pick |
| 3 | 13 | liquid |
| 4 | 13 | colony pick |
DNA Extraction purgene kit
- 5ul of overnight culture was spread on a plate with chloramphenicol to have a stock of the bacteria tested and grown overnight at 37C
- Heat block warmed to 37C
- Cell lysis solution chilled on ice until cloudy
- 500ul of overnight solution per tube was added to a 1.5mL tube
- Tubes were spun down for 30 seconds at 13,000rpm
- Supernatant was discarded
- Added 300ul of chilled cell lysis solution
- Added 0.6ul of 10mg/mL RNase A and inverted 25 times and spun down
- Place tubes on heat block at 37C for 1 hour
- Prepared fresh 70% ethanol and final 1.5mL tubes with 350ul of 100% isopropanol
- After incubation, placed tubes on ice
- Added 150ul of protein precipitation solution
- Vortexed tubes and placed on ice for 5 minutes
- Centrifuged tubes for 5 min at max speed
- Transferred supernatant (~450ul) to the final tubes with isopropanol and inverted 50X
- Centrifuged tubes for 3 min at max speed
- Discarded supernatant
- Added 300ul fresh 70% ethanol and inverted twice
- Centrifuged tubes 1 minute max speed
- Discarded supernatant
- Let tubes air dry on kim wipe ~30 min
- Resuspended pellet in 100ul of DNA hydration solution - DNA amount was large and visible
p47 PCR and gel
- Thawed reagents and primer on ice and vortexed and spun down
- Made master mix on ice:
- 32.5ul GoTaq
- 1.625ul F primer
- 1.625ul R primer
- 19.5ul molecular grade water
- Master mix vortexed and spun down before use
- Added 9ul of mix to strip tubes
- Added 1ul of DNA to respective tube
- Added 1ul of molecular grade water to the negative control
- Used a positive control
- Vortexed and spun down strip tubes before placing in the p47 PCR program for 35 cycles
After ran on a 1% gel for 35 min at 90V. The first six wells are the only ones to look at in this gel, all are negative for p47 except for the positive control. These colonies were not investigated further. Information on colonies screened is here