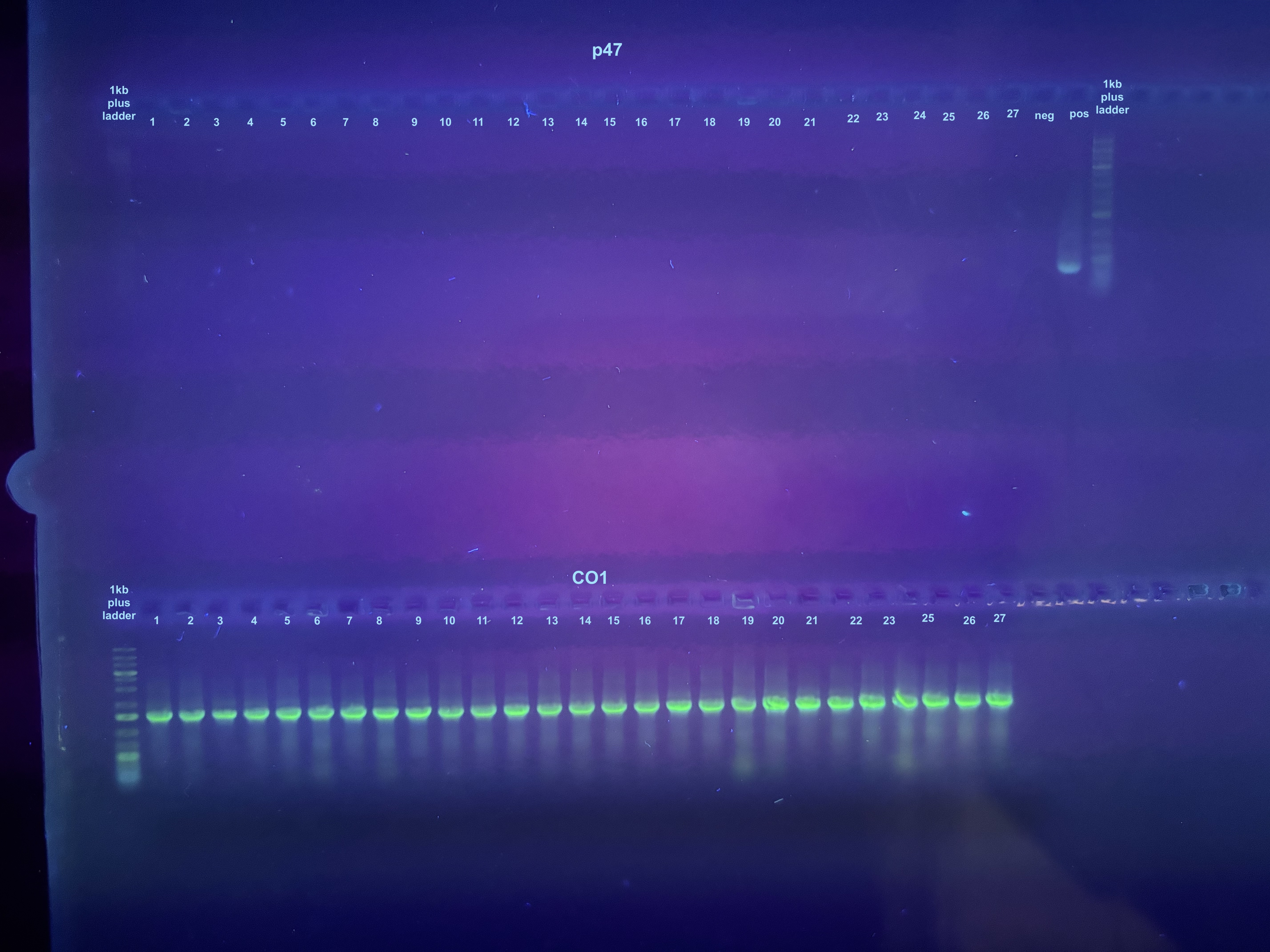
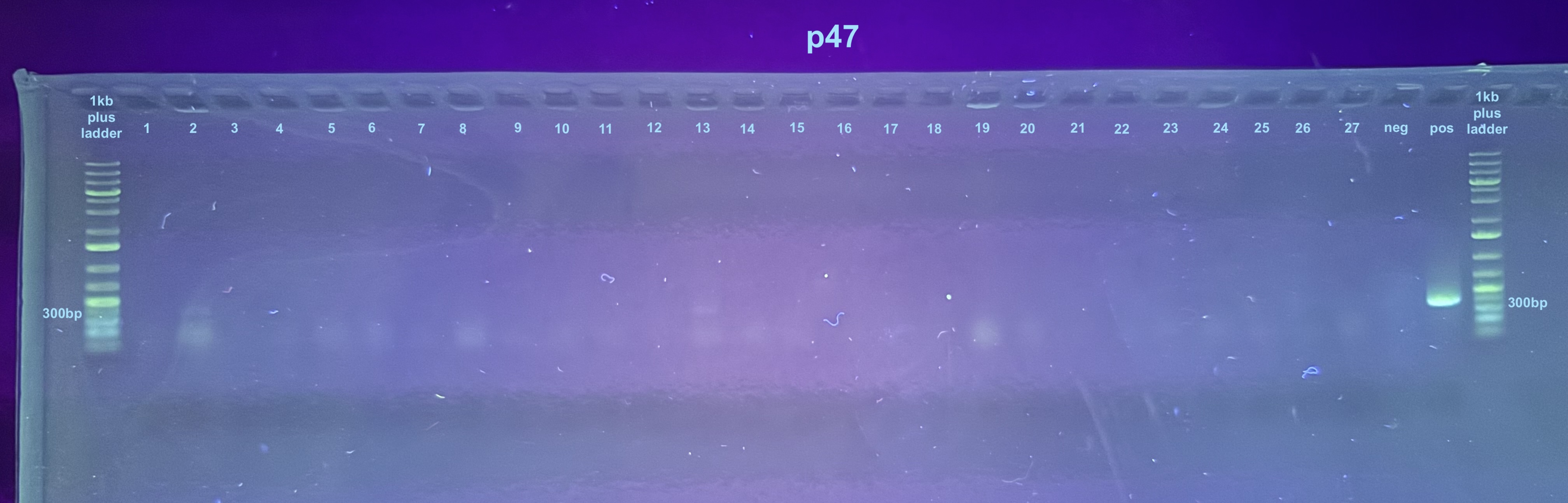
Trying Colony PCR on Plates from Electroporation of pSPIN-BAC E. coli with DiNV DNA
I decided to pick 3 colonies of each of the plates prepared after electroporation and attempt colony PCR on them. I used this Addgene blog for help on how to plan the PCRs. Rob suggested doing a positive control PCR and using 16S (bacterial) to see if the colony PCR works at all.
All of the plates from the day before had bacterial growth so it at least seems like the preparation of electrocompetent cells did not kill all the bacteria.
I prepared 27 strip tubes with 6ul of molecular grade water in them, although when I was picking the colonies I felt like the bacteria I picked up was such a small amount that 6ul would be too much, so for most of the samples I removed 2ul so it should be ~4ul each colony was put in.
For each colony, I picked it with an autoclaved pipette tip and swirled it in the tube of water for a few seconds:
| colony/tube # | plate | type | volume water |
|---|---|---|---|
| 1 | 2 | Neg 1 | 6ul |
| 2 | 2 | Neg 1 | 6ul |
| 3 | 2 | Neg 1 | 6ul |
| 4 | 4 | Neg 2 | 6ul |
| 5 | 4 | Neg 2 | 4ul |
| 6 | 4 | Neg 2 | 4ul |
| 7 | 7 | DiNV | 4ul |
| 8 | 7 | DiNV | 4ul |
| 9 | 7 | DiNV | 4ul |
| 10 | 8 | DiNV | 4ul |
| 11 | 8 | DiNV | 4ul |
| 12 | 8 | DiNV | 4ul |
| 13 | 9 | DiNV | 4ul |
| 14 | 9 | DiNV | 4ul |
| 15 | 9 | DiNV | 4ul |
| 16 | 10 | DiNV | 4ul |
| 17 | 10 | DiNV | 4ul |
| 18 | 10 | DiNV | 4ul |
| 19 | 11 | DiNV | 4ul |
| 20 | 11 | DiNV | 4ul |
| 21 | 11 | DiNV | 4ul |
| 22 | 12 | DiNV | 4ul |
| 23 | 12 | DiNV | 4ul |
| 24 | 12 | DiNV | 4ul |
| 25 | 13 | DiNV | 4ul |
| 26 | 13 | DiNV | 4ul |
| 27 | 13 | DiNV | 4ul |
- Prepared 10uM 16S primers:
- 90ul of molecular grade water
- 10ul of primer
- Vortexed and spun down and kept ice
- Thawed other reagents and primers on ice, vortexed and spun down
- Prepared master mixes on ice:
| reagent | 16S volume | p47 volume |
|---|---|---|
| GoTaq | 150ul | 150ul |
| Forward primer | 7.5ul | 7.5ul |
| Reverse primer | 7.5ul | 7.5ul |
| molecular grade water | 105ul | 105ul |
- Vortexed and spun down master mixes
- Aliquoited 9ul of master mix to strip tubes
- Added 1ul of colony-water mix to each tube
- Added 1ul of molecular grade water, and 1ul of positive DNA for the p47 primers to their respective tubes
- Vortexed and spun down
- Placed tubes in the p47 and 16S PCR programs, each cycling 35 times
Ran 2 gels, the first gel did not show the p47 samples because of a dye issue so I ran it again separately. So far no real evidence of retained DiNV DNA…