Second Exonuclease V Treatment Test on Hirt Extracts to Attemp to Remove Linear DNA and 6 Hour Gel
Because the gel of the previous treatment test was really hard to see for one of the crucial samples, I decided to try the treatment and cleanups again on just the cell pellet samples. Sample information can be found here
20240118 Exonuclease Treatment
- Followed exact same process as the first use, where the protocol was based off of this protocol
- I used the rest of the DNA in the 16 and 17 samples to try to maximize what I could
| sample | DNA | 10mM Tris to 30ul | total DNA |
|---|---|---|---|
| 16 | 16ul | 14ul | ~900ng |
| 17 | 16ul | 14ul | ~900ng |
- All DNA was kept on ice, flicked to mix, and spun down before use
- All reagents were thawed on ice, vortexed to mix, and spun down before use
- The enzyme was not frozen, and was spun down before use
- DNA was diluted into 1.5mL tubes
- To each diluted sample tube:
- Added 4ul of NEBuffer 4
- Added 4ul of ATP
- Added 2ul of exonuclease V
- Tubes were flicked to mix and spun down
- Tubes were placed in the 37C heat block for 1 hour
- Then tubes were placed in a 70C heat block for 30 minutes to inactivate the enzyme
- Samples were kept on ice while awaiting cleanup
I still wanted to see if there was a difference in size retained from the NEB Monarch cleanup or a phenol chloroform cleanup, so I tried both.
NEB Monarch DNA Cleanup
- Using sample 17 for this
- Using NEB Monarch PCR and DNA kit protocol
- Warmed DNA elution buffer to 50C in heatblock
- Increased the volume in the tube from 40ul to 200ul with 160ul of 10mM tris
- Added a 2:1 ratio of DNA binding buffer to the tube: 400ul each
- Inverted tube to mix and spun down
- Added the total volume (~600) of sample to a spin column
- Centrifuged the column at 16,000rcf for 1 minute
- Discarded the flow through
- Added 200ul DNA wash buffer to the column
- Centrifuged the column at 16,000rcf for 1 minute
- Discarded the flow through
- Added another 200ul DNA wash buffer to the column
- Centrifuged the column at 16,000rcf for 1 minute
- Discarded the flow through
- Centrifuged the column at 16,000rcf for 1 minute “dry”
- Transferred the column to new labeled 1.5mL tubes
- Added 15ul of warmed DNA elution buffer to the center of the filter and waited 2 minutes
- Centrifuged the column at 16,000rcf for 1 minute
- Placed the sample in the 4C for storage
Phenol-Chloroform Cleanup
- Using sample 16
- Basing protocol off of what I’ve done in the hirt extractions (see example and this document)
- All work was done in the fume hood
- Increased volume in sample to 300ul with 260ul of 10mM tris
- Added equal volume (~300ul) of cold phenol-chloroform isoamy alcohol
- Inverted to mix
- Placed tube on orbital shaker for 10 minutes
- Centrifuged tube for 15 minutes at 16,000rcf at room temp
- Removed the top layer into new final labeled tubes:
- 16: 300ul
- Added 0.1X volume (30ul) of 3M sodium acetate to the tube
- Added 2-2.5X volume of cold 100% ethanol to the tube: I added 750ul which is 2.5X
- Mixed by inverting many times
- Placed sample in the -20 overnight
- Placed the centrifuge in the 4C to cool overnight
- Next day 20240118
- Centrifuged tube at 4C for 30 minutes at 16,000rcf
- I did saw a very small pellet
- Poured off the supernatant into the waste
- Added 500ul of cold fresh 80% ethanol
- Inverted the tube twice
- Centrifuged the tube at 4C for 30 minutes at 16,000rcf
- Poured off the supernatant into the waste
- Let the tube dry for ~10 minutes upside down
- Resuspended the pellet in 15ul of 10mM tris
- Let pellet resuspend for about 10 minutes before use in the gel
20240118 Qubit
- High sensitivity Qubit of the exonuclease treated samples, there may not really be anything left in these
- Followed this Qubit protocol
- 16: 8.78ng/ul
- 17: 2.22ng/ul
20240118 6 Hour gel
- Prepared a 0.7% gel with the Seakem agarose that is supposed to be better for gels with HMW DNA
- 160mL of 1X TAE
- 1.155g Seakem agarose
- Microwaved until clear solution
- Added 3ul of Midori stain and swirled
- Saved 1mL in a 1.5mL tube and placed in the heat block at 65C to stay warm and liquid
- Poured the large gel and waited for it to solidify
- Once solid, added 1 round of PFG marker to the 1st and 8th wells
- Let the tube of gel cool for 2-3 minutes
- Filled the wells with the PFG markers with the extra agarose and waited for those to solidify
- Prepared the samples:
- 16-exo: 14ul of DNA and 2.8ul of dye
- 17-exo: 14ul of DNA and 2.8ul of dye
- Added 48kb ladder and samples into the gel
- Set the gel for 58V and 6 hours
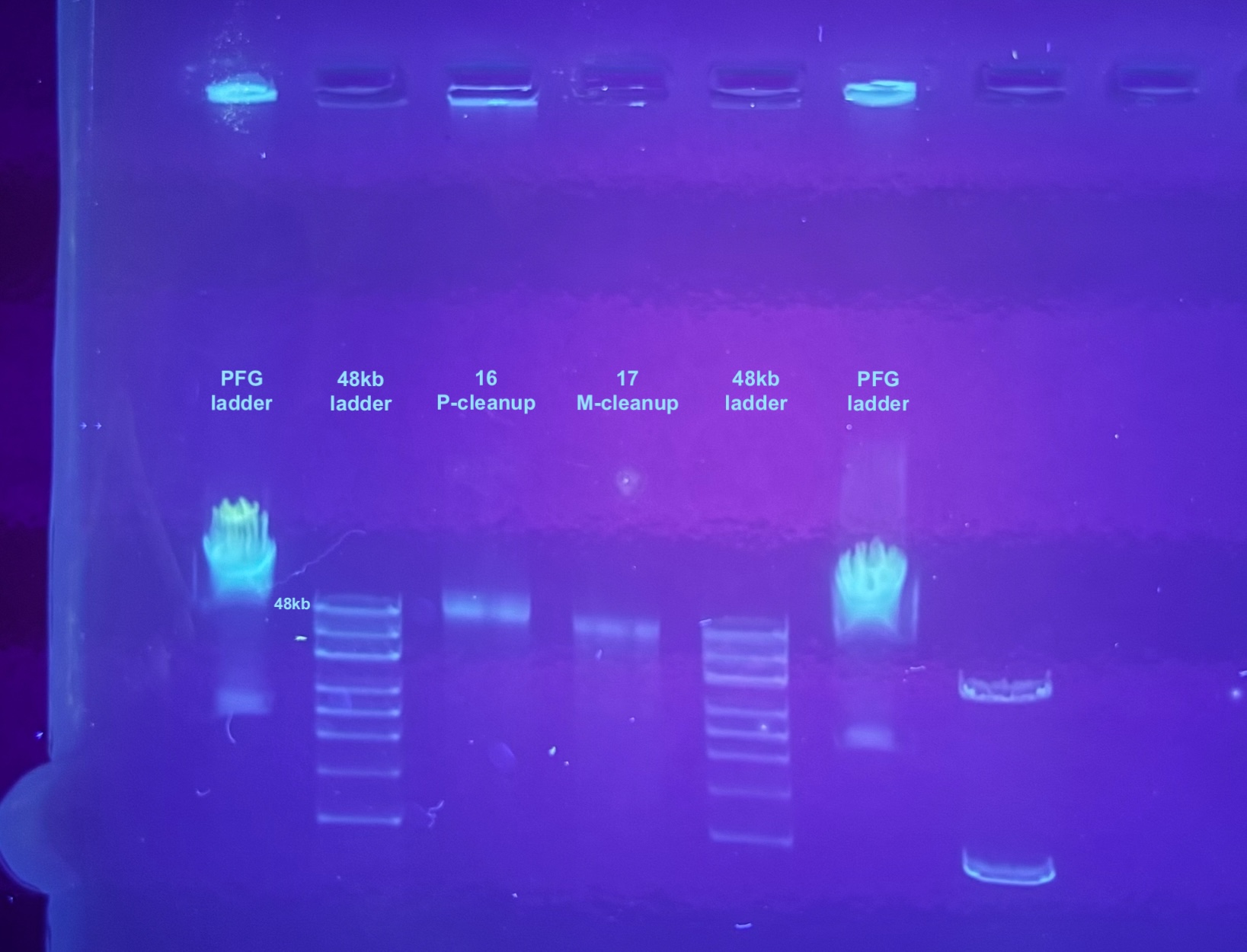
This looks like the phenol cleanup could yield slightly higher molecular weight DNA, although I am not sure if this indicates intact viral genomes or not, it seems a little small. But I know that after ~50kb they just don’t run very well on the gel. I also found some information online that suggests supercoiled closed circular DNA will run faster in a gel than you’d expect based on size. And this would be the form left from the exonuclease? I think it is best to proceed with using the phenol cleanup method after this treatment in the future.