Testing Conditions for qPCR When doing Adult Fly DiNV Infection Experiments
Previous qPCR of adult flies tested different dilutions of DNA failed to indicate a good dilution of DNA to use for these experiments. We are running into the issue where some control samples come up with a small amount of fly or virus DNA, and there isn’t a good dilution that removes that. That experiment only looked at day0 flies, where the amount of DiNV was very low. For this test we included more controls and also flies who died from the infection. We haven’t looked at the qPCR results of flies dead from infection yet.
Samples to test
| sample | Bio reps | dilution1 | dilution2 | primers | Tech reps | Total wells |
|---|---|---|---|---|---|---|
| Day0 CCM | 2 | 1ng | 0.5ng | TPI and PIF3 | 3 | 24 |
| Day0 DiNV injection | 2 | 1ng | 0.5ng | TPI and PIF3 | 3 | 24 |
| Extraction control | 2 | Not diluted | NA | TPI and PIF3 | 3 | 12 |
| Dead from DiNV injection | 2 | 1ng | 0.5ng | TPI and PIF3 | 3 | 24 |
| water | NA | none | NA | TPI and PIF3 | 6 | 12 |
These samples were DNA extracted and diluted to these specific concentrations here. The extraction controls used in this qPCR are from that DNA extraction.
Plate layout for the experiment:
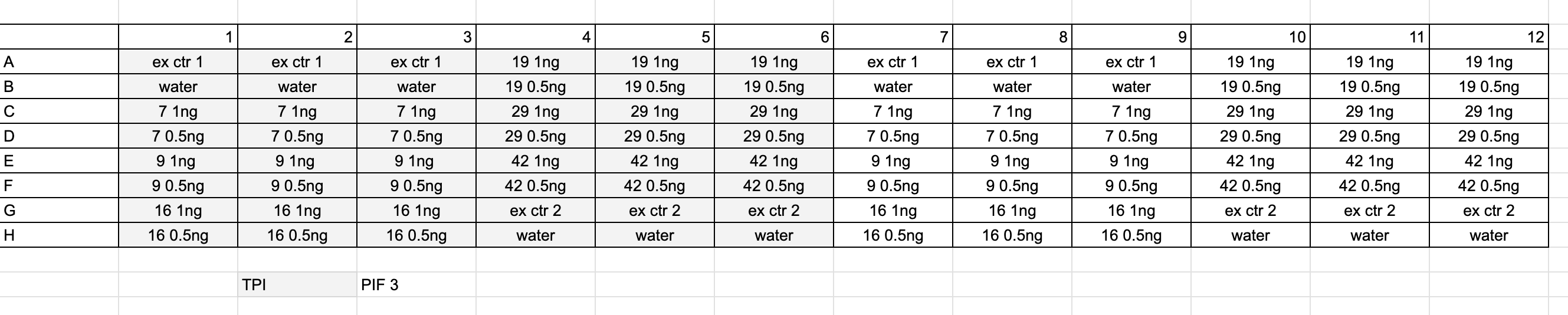
- Samples and reagents were thawed on ice, vortexed, and spun down before use
- There were 48 individual wells for each primer, 3 were added on for pipetting error
- PIF3 master mix:
- 5ul Sso supermix * 51 = 255ul
- 0.5ul PIF3 F primer * 51 = 25.5ul
- 0.5ul PIF3 R primer * 51 = 25.5ul
- 3ul nuclease free water * 51 = 153ul
- The master mix was made on ice, vortexed, and spun down
- TPI master mix:
- 5ul Sso supermix * 51 = 255ul
- 0.5ul TPI F primer * 51 = 25.5ul
- 0.5ul TPI R primer * 51 = 25.5ul
- 3ul nuclease free water * 51 = 25.5ul
- The master mix was made on ice, vortexed, and spun down
- I used the qPCR specific plates and seals
- 9ul of the appropriate master mix was added to the planned well (see layout above)
- 1ul of DNA was added to the planned well (see table above)
- Each well was pipette mixed with 5ul using a multichannel
- The plate was sealed and centrifuged for 6 minutes at 3,000g
- The plate was put in the qPCR machine:
- Used program CFX manager
- Selected user-defined protocol, existing protocol, and KMM folder
- Selected the p47 program
- Used the machine buttons to open and close the lid
- Started the program
- Data from the qPCR machine can be found here
- And analysis of the Cq results can be found here as a markdown