Checking Myd88 Samples with RP49 and Dpt PCR Primers
The previous PCRs on Myd88 cells and supernatant DNA had some hazy/faint bands for the RP49 primers, so Rob wanted me to try the Dpt cw (diptericin) primers on some samples. I picked 3 samples of Myd88 cells on day 0 because I could use their DNA and not waste it because I’m not using those samples for further qPCR. However this might not have been the best idea because the DNA concentration is probably very low in these so the gel didn’t end up showing much.
For a positive control I used sample 16 from the first attempt at the Myd88 DiNV infection experiment, it is a Myd88 sample.
- DNA and reagents were thawed on ice, vortexed, and spun down before use
- RP49 master mix:
- 5ul GoTaq * 6 = 30ul
- 0.25ul RP49 F * 6 = 1.5ul
- 0.25ul RP49 R * 6 = 1.5ul
- 3.5ul nuclease free water * 6 = 21ul
- The master mixed was vortexed, spun down, and kept on ice
- Dpt master mix:
- 5ul GoTaq * 6 = 30ul
- 0.25ul Dpt F * 6 = 1.5ul
- 0.25ul Dpt R * 6 = 1.5ul
- 3.5ul nuclease free water * 6 = 21ul
- The master mixed was vortexed, spun down, and kept on ice
- Reaction tubes were set up on ice
- 9ul of master mix was added to the appropriate strip tubes
- 1ul of DNA was added to the appropriate strip tubes
- 1ul of positive control was added to the positive control tubes
- 1ul of nuclease free water was added to the negative control tubes
- Strip tubes were vortexed and spun down
- The Dpt tubes were put in Sarah’s Dpt40 program with cycles reduced to 35 (annealing temp is 57.5C)
- The RP49 tubes were put on the 115 PCR program (this program also might not be the best for these primers)
- After PCR, a 2% gel was run for 35 minutes
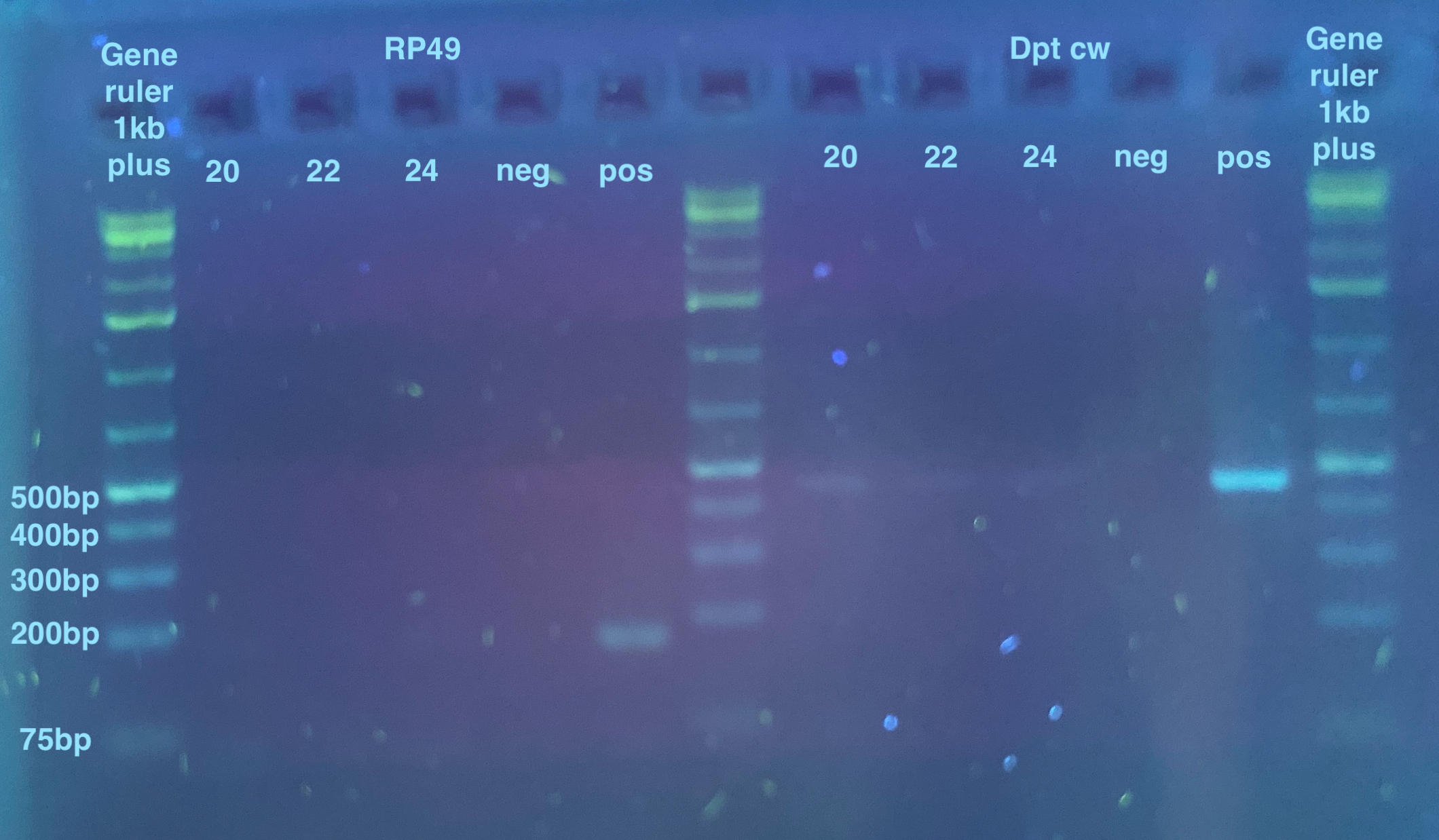
The bands are very faint here, but I can see bands for all the Dpt samples, so I felt confident that these samples have melanogaster DNA in them, and probably all the other RP49 faint ones too.