PCR on RPL11 and p47 on the 3mLC Gel Extractions, and Testing the GoTaq
20220104 RPL11 with NEB Taq on 3mLC gel extraction samples
- Based off of Qubits from the extractions, I will use 1-2ul of DNA per reactions (~7-10ng per reaction)
- Made master mix on ice:
- 1ul NEB buffer * 6.6 = 6.6ul
- 1ul dNTPs * 6.6 = 6.6ul
- 1ul MgCL2 * 6.6 = 6.6ul
- 0.25ul vir_RPL11_F * 6.6 = 1.65ul
- 0.25ul vir_RPL11_R * 6.6 = 1.65ul
- 0.1ul NEB Taq * 6.6 = 0.66ul
- 4.4ul molecular grade water * 6.6 = 29.04ul
- Mix was vortexed and spun down
- The positive control is the 3mL HMW extraction sample, diluted to ~10ng/ul
- Strip tubes were made and mix and sample were added to them:
| tube # | sample | ul MM | ul DNA | ul water |
|---|---|---|---|---|
| 1 | 3mLC-150 | 8 | 2 | 0 |
| 2 | 3mLC-UP | 8 | 1 | 1 |
| 3 | 3mLC-LOW | 8 | 2 | 0 |
| 4 | 3mLC-ADD | 8 | 1 | 1 |
| 5 | Neg control | 8 | 0 | 2 |
| 6 | Pos control | 8 | 1 | 1 |
- Samples were mixed and spun down and placed in the PCR program:
- 95 degrees C 3 minutes
- 95 degrees C 30 seconds
- 57 degrees C 1 minute
- 68 degrees C 1 minute 30 seconds
- 68 degrees C 5 minutes
- 12 degree C hold
- bold sections are cycled 34 times
- The PCR was ran on a 2% gel for 30 minutes at 90V the next day:
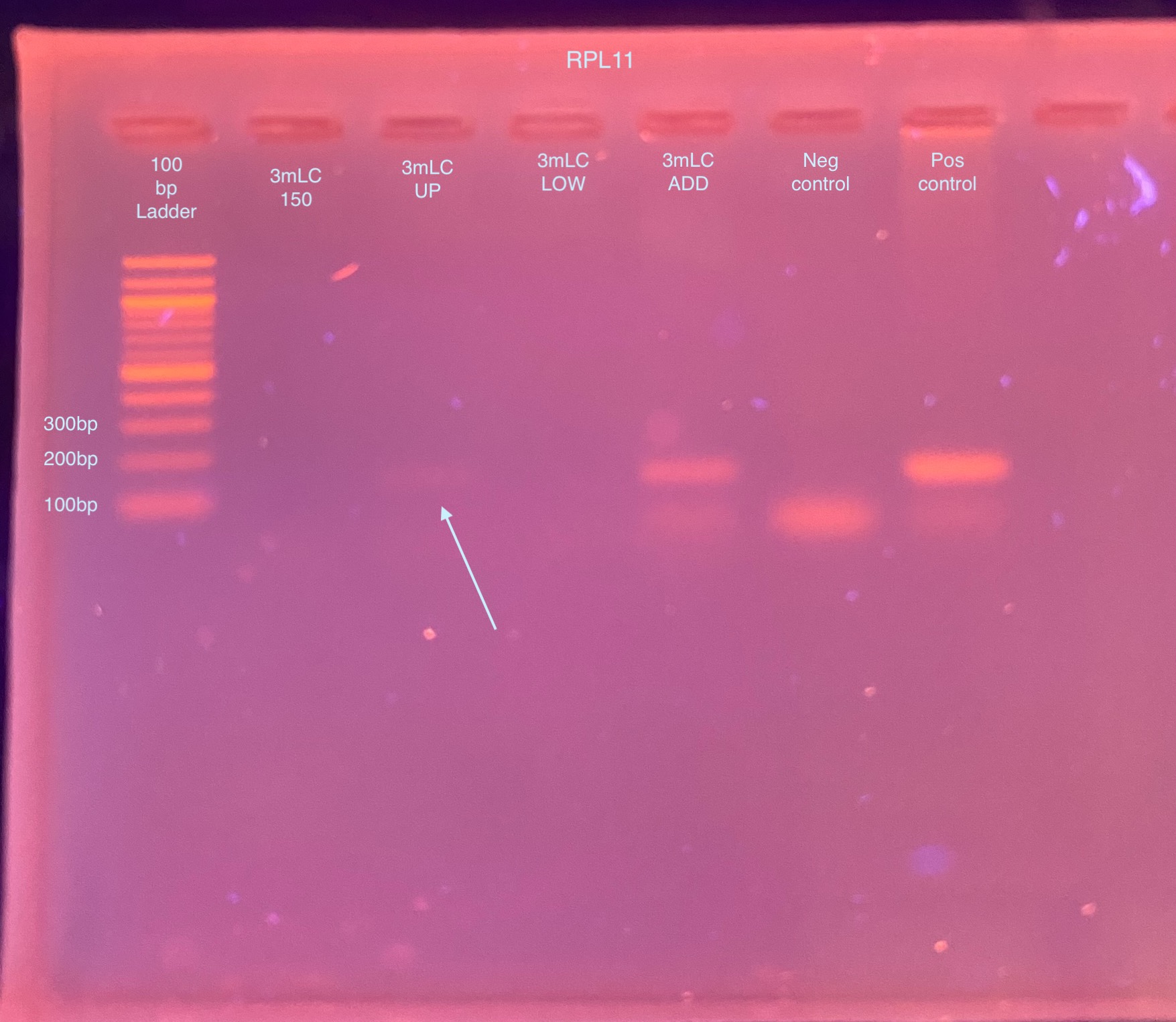
- This was somewhat promising because for once I got amplification in 1 (maybe 2) of the gel extractions! I decided to try the same PCR with the GoTaq from Jessie
20220106 RPL11 with GoTaq on 3mLC gel extraction samples
- Made master mix on ice:
- 5ul GoTaq * 6.6 = 33ul
- 0.25ul vir_RPL11_F * 6.6 = 1.65ul
- 0.25ul vir_RPL11_R * 6.6 = 1.65ul
- 3.5ul molecular grade water * 6.6 = 32.1ul
- Mix was vortexed, and kept on ice
- 9ul of master mix was added to each of 6 strip tubes
- 1ul of DNA was added to each strip tube (in order above), except the negative control, which got 1ul molecular grade water
- Tubes were vortexed, spun down, and placed in the RPL11 PCR program
- A 2% gel was ran the next day for 30 minutes at 90V:
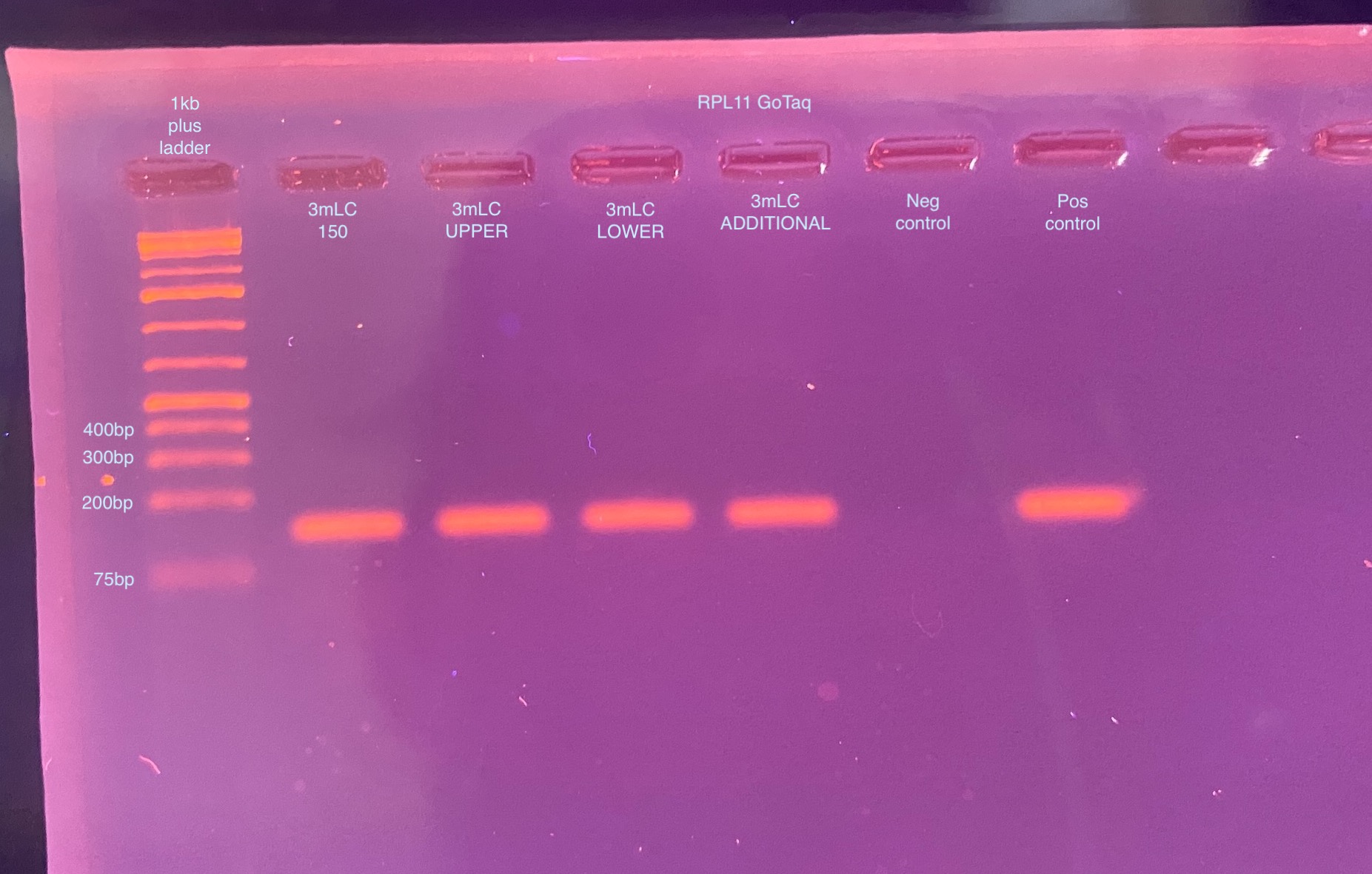
- Wow the GoTaq looks great! Every sample amplified really well, and some of them had lower DNA inputs!
20220107 p47 PCR with GoTaq on 3mLC gel extraction samples
- Made master mix on ice:
- 5ul GoTaq * 6.6 = 33ul
- 0.25ul p47_F * 6.6 = 1.65ul
- 0.25ul p47_R * 6.6 = 1.65ul
- 3.5ul molecular grade water * 6.6 = 32.1ul
- Mix was vortexed, and kept on ice
- 9ul of master mix was added to each of 6 strip tubes
- 1ul of DNA was added to each strip tube (in order above), except the negative control, which got 1ul molecular grade water
- Tubes were vortexed, spun down, and placed in the p47 PCR program:
- 95 degrees C 5 minutes
- 95 degrees C 30 seconds
- 55 degrees C 30 seconds
- 72 degrees C 30 seconds
- 72 degrees C 5 minutes
- Hold at 12 degrees C
- bold text is cycled through 30 times
- A 1% gel was ran the next week for 50 minutes at 90V
- Additionally, I tried adding the non-PCR’d gel extraction DNA for one of the sample on after the positive control, but it completely floated out of the well. So, I’m not sure why that is/what is happening. Clearly it’s ok to PCR, at least with the really robust GoTaq. If it is ethanol, I don’t really know how to get rid of it
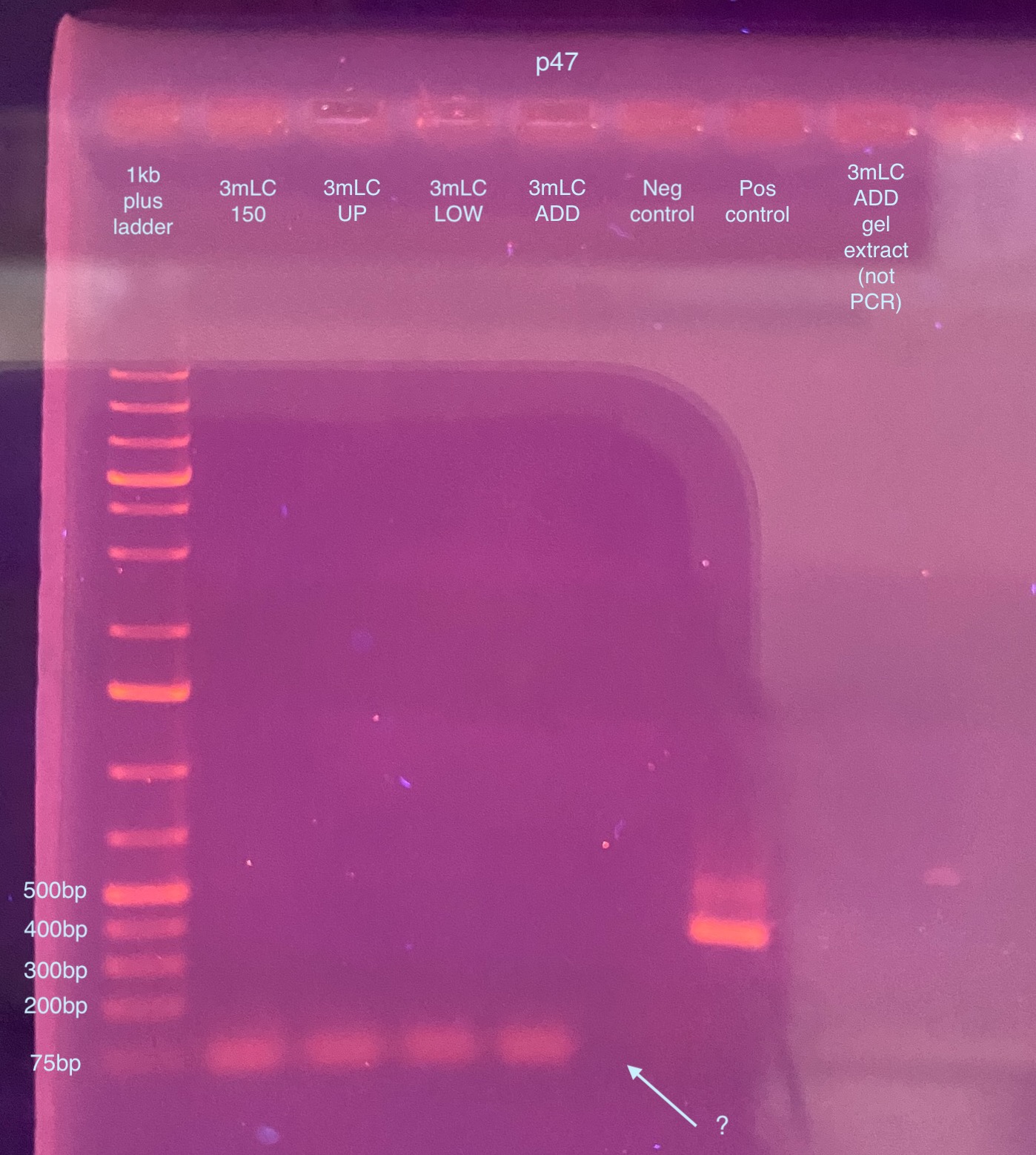
- This looks like there’s no p47 amplification for any sample, not good. However it is weird that there’s no primer dimer for the negative control. So I decided to try this PCR again just in case something was off. You also can’t see anything for the DNA I added for the non - PCR’d gel extraction sample
20220111 p47 PCR again with GoTaq on 3mLC gel extraction samples
- Followed exactly the same thing as above, but I made a new aliquot of molecular grade water in case that was somehow causing a problem (however that same water is in the master mix so it doesn’t seem like that is the issue)
-
- Made master mix on ice:
- 5ul GoTaq * 6.6 = 33ul
- 0.25ul p47_F * 6.6 = 1.65ul
- 0.25ul p47_R * 6.6 = 1.65ul
- 3.5ul molecular grade water * 6.6 = 32.1ul
- Mix was vortexed, and kept on ice
- 9ul of master mix was added to each of 6 strip tubes
- 1ul of DNA was added to each strip tube (in order above), except the negative control, which got 1ul molecular grade water
- Tubes were vortexed, spun down, and placed in the p47 PCR program:
- 95 degrees C 5 minutes
- 95 degrees C 30 seconds
- 55 degrees C 1 min
- 72 degrees C 1 min 30 seconds
- 72 degrees C 5 minutes
- Hold at 12 degrees C
- bold text is cycled through 30 times
- A 1% gel was ran that day for 50 minutes at 90V
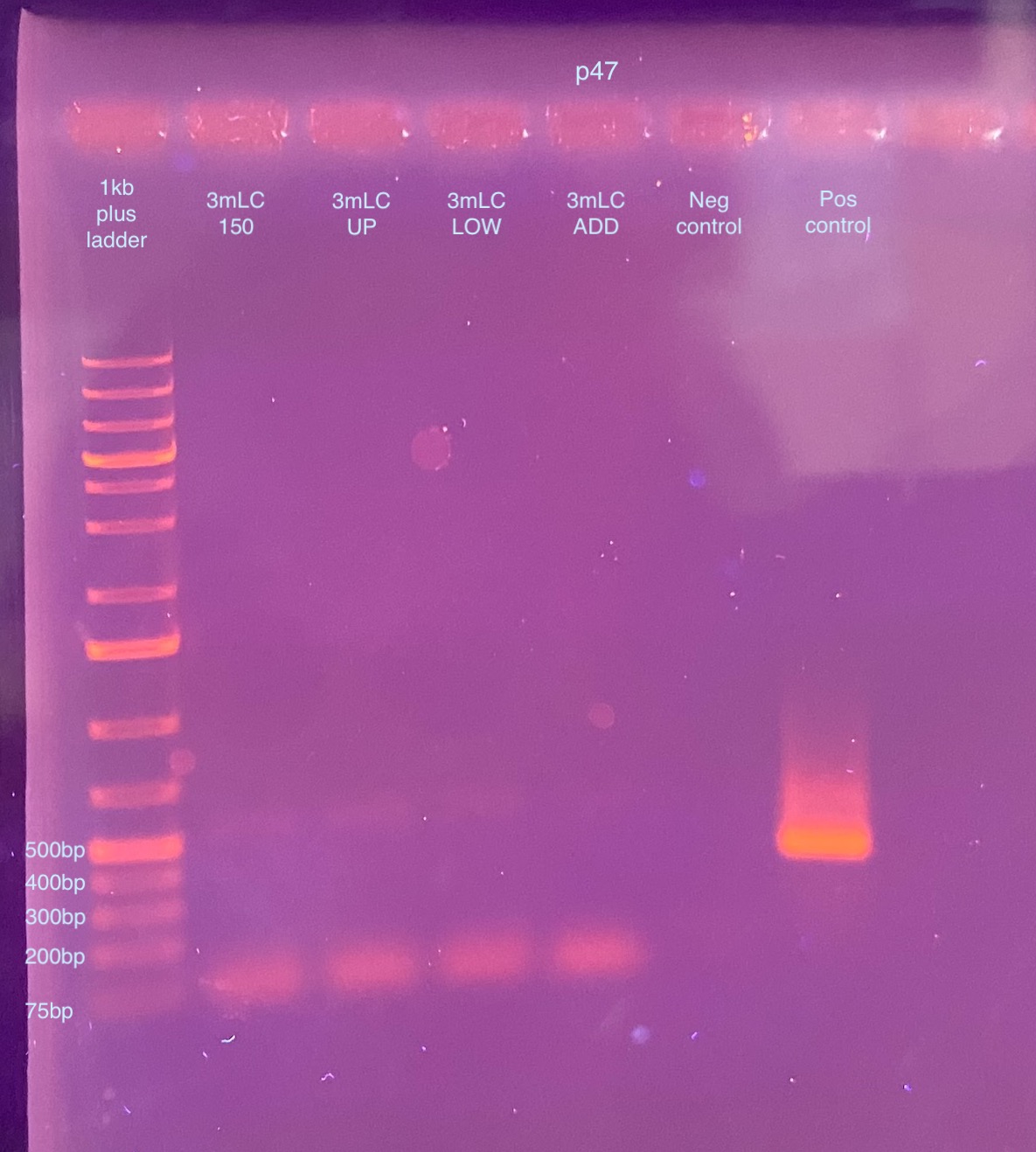
- This gel looks exactly the same. I went back through a lot of my gel images and I don’t often see primer dimer show up in the images (so small usually) so this is getting me worried that the band in the gel extraction samples might actually be DNA amplifying. And that it’s so crushed up somehow from the gel extraction procedure that it can only amplify a small piece? I looked up online and primer dimer is usually 20-50bp size. This seems to be 75bp or maybe a little larger? I’m not sure what is going on. I can’t see the DNA on the gel because it all floats out, so I have no idea the size.