Another Overnight Gel and Zymoclean Large Fragment DNA Recovery Kit on 3mL-120hr HMW DNA Sample, Using Recommendations From Zymo Representatives
Notes
- Using 3mL-120hr HMW DNA sample (~115ng/ul)
- Skipped wells in the gel when loading to give ample room for slicing
- Minimized time on UV,
- I implemented all modifications to the Zymo kit to maximize DNA yield and length:
- Heated elution buffer to 70 degrees C
- Did two elution centrifuges
- Incubated elution liquid on the column for >1 minute each time (5 min each)
- Additionally I added a “dry spin” after the 2nd wash for 1 minute to dry the column of any residual wash buffer (ethanol)
- From the Zymo Rep the suggested:
- Incubating the gel slice while it dissolves for ~20min or until there were no swirls when tapping the tube
- Increasing the centrifugation speed to 16,000rcf
20220103 Loading and Running Gel
- 0.8% gel mix:
- 180mL 1X TAE
- 1.4g agarose
- Microwaved for ~4min
- Pipetted 1mL into a 1.5mL tube and saved on the heat block at 65 degrees C make sure the heat block is on and running
- Made sure the large tray had well sealed tape barriers
- Let the gel liquid cool for ~10 min until pouring into the tray and placing in the combs
- Gel cooled in ~20 min or less
- Added 1 round of PFG ladder to the left-most well
- Let the 1mL saved gel cool for ~3 min outside of the heat block
- Filled the PFG well with cooled gel liquid to the top of the well to seal the round in
- Waited ~3 minutes for the wells to cool
- Made up 48kb ladder:
- 1.2ul ladder
- 6ul loading dye
- 28.8ul molecular grade water
- Placed the gel tray into the box after removing the tape
- Mixed 2ul of loading dye with 10ul of 3mL-120hr sample (with a clipped tip) and added it to the 3rd well in the gel
- Added 16ul 48kb ladder to the 5th well in the gel
- Set the timer for 16.5 hours at 40 volts and started it at ~5:02pm
20211208 Gel Slicing and Extraction
- Stopped the gel at 9am
- Sliced the gel in half and placed the half with samples in the EtBr bath for 1 hour
- While that was in the bath, I made up final tubes and weighed tubes for the starting weight for the gel extraction
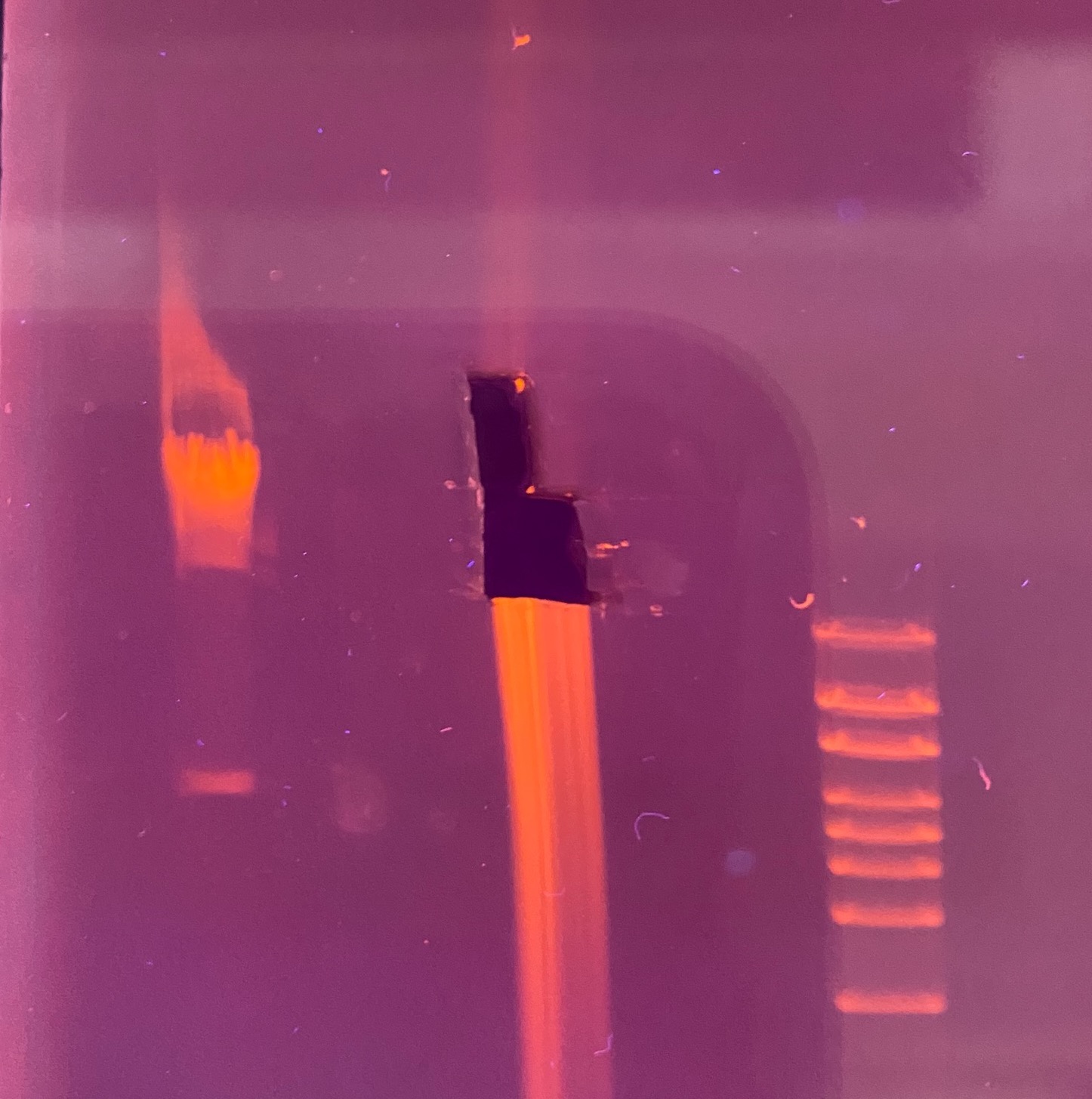
- After 1 hour: put on PPE for looking at UV and took picture of the gel
- Here I noticed the gel was a little strange (see image below), there is an upward tail of DNA that either is that large or ran funny that’s above the usual huge clump of HMW DNA. I decided to slice out that piece as a separate piece (“additional”), so I quick made up another tube for it and made measurements
- Cleaned fresh razor blade with ethanol before use
- Quickly sliced out a square of the whole “HMW” section of the 3mL-120hr sample, then made quick slices to cut the lower, middle, and upper pieces
- Then slice out the top additional section
- Turned off the UV for manipulating the pieces out of the gel and into their tubes
- Gel image and approximate slice positions:

- Looks like the gel ran kind of off angle again, because the 48.5kb positions in the ladders don’t line up, but if it did just run slightly off, then what I sliced should have the 150kb piece in it somewhere
- Here is the gel without the slices in it:
- Cleaned up and disposed properly all EtBr touching items
- Weighted every sample tube with the gel in it, and calculated the gel weight, and the 3X gel weight
| tube name | tube weight (mg) | tube weight with gel (mg) | gel weight (mg) | 3X gel weight volume (ul) |
|---|---|---|---|---|
| 3mLC-150 | 984.1 | 1032.4 | 48.3 | 144.9 |
| 3mLC-UP | 987 | 1046.4 | 59.4 | 178.2 |
| 3mLC-LOW | 987.3 | 1013 | 25.7 | 77.1 |
| 3mLC-ADD | 994.3 | 1077 | 82.7 | 248.1 |
- Added 3 volumes of gel weight (5th column in table)of ADB to each sample tube (kit buffer)
- Incubated sample tubes at 50 degrees C in the heat block for 20 minutes, then checked and most tubes still had a swirl in them, so they were let go for another 5 minutes. After this, all tubes but 3mLC-ADD were clearly dissolved. I kept that tube in the heat block until I had finished adding the other tubes to their spin columns
- After this, the heat block was bumped to 70 degrees and elution buffer was incubated until later
- Clipped tips of pipettes and added the total volume of each dissolved gel tube to individual spin columns and collection tubes provided by the kit
- Centrifuged spin columns for 1 minute at 16,000rcf
- Pipetted out flow through
- Added 200ul of DNA wash buffer to each column
- Centrifuged columns for 30 seconds at 16,000rcf
- Pipetted out flow through
- Added another 200ul DNA wash buffer to each column
- Centrifuged for 30 seconds at 16,000rcf
- Pipetted out flow through
- Centrifuged for 30 seconds at 16,000rcf “dry”
- Transferred columns to final 1.5mL tubes
- Added 15ul of 70 degree C DNA elution buffer directly to the filters in the columns
- Incubated the columns on the bench for 5 minutes
- Centrifuged the columns for 30 seconds at 16,000rcf
- Added 15ul of 70 degree C DNA elution buffer directly to the filters in the columns
- Incubated the columns on the bench for 5 minutes
- Centrifuged the columns for 30 seconds at 16,000rcf
- Put the tubes on ice for Qubiting
- Qubit HS DNA Assay:
- 3mLC-150: 5.12ng/ul
- 3mLC-UP: 7.42ng/ul
- 3mLC-LOW: 2.14ng/ul
- 3mLC-ADD: 8.13ng/ul