PCR of p47 (DiNV) and COI (control) on Gel Extraction Products from Zymoclean Gel Extraction Determine if Virus DNA is in the HMW Fractions
Notes
- Decided to add MgCl2 to the PCR reactions because I think other people use it. Also the p47 positive control in the last PCR I did was pretty faint, so I’m hoping this will help that. I took an aliquot from Kent’s box for my own. It’s 25mM in concentration. I couldn’t find any protocols in our lab drive with the desired concentration of MgCl2, but googling online suggests 1.5-4.5mM concentration in a reaction for it. So if I use it at 1ul per reaction in a 10ul reaction, that would be 2.5mM
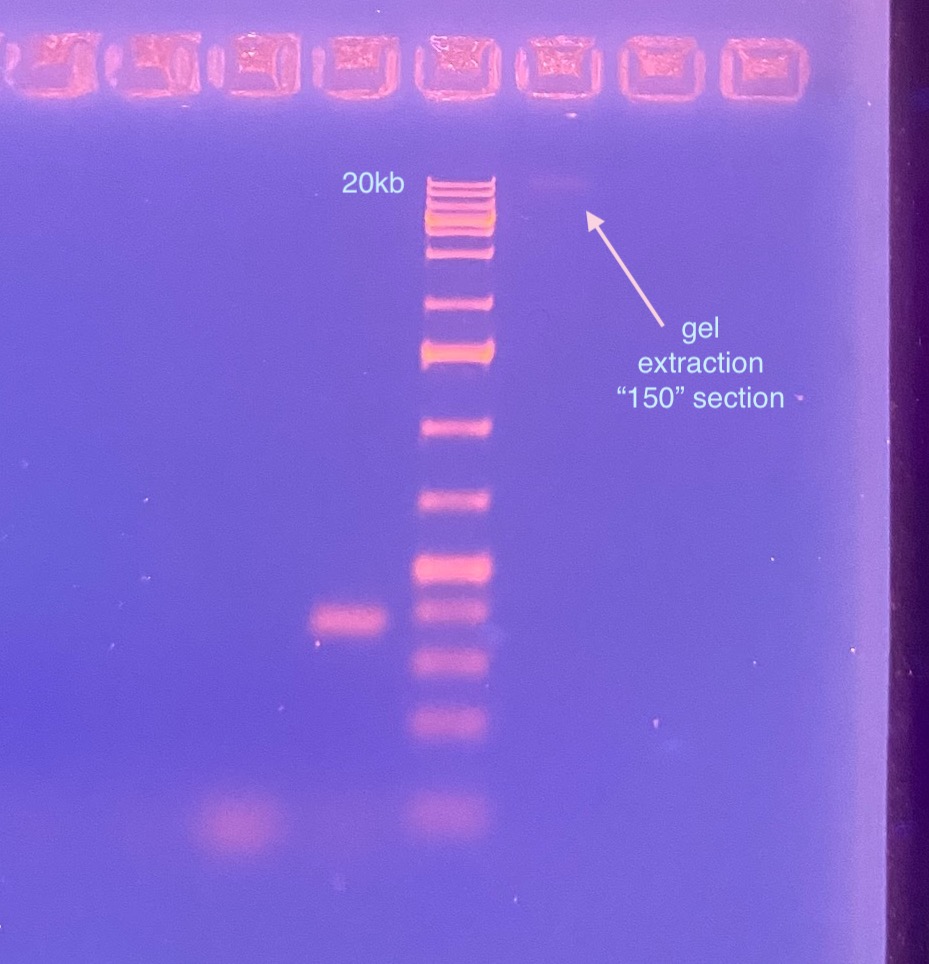
- I decided to gel the “150” slice gel extract to see if it would show up on the PCR product gel, and to see if it would be HMW still
202111201 COI PCR
- Planned to run all samples from the extraction, even the gel control and the PFG, plus a negative and positive control, equalling 8 samples, plus 0.8 for error. So I made enough mix for “8.8”
- The positive control is a from Kent and is a positive control for p47 (virus)
- All samples (except for the positive control) have somewhat low DNA, so I used an input volume of 2ul for the DNA samples (and 1ul for the positive control)
- All samples were kept on ice, along with primers, and enzymes
- Made master mix on ice:
- 1ul 10X NEB taq buffer * 8.8 = 8.8ul
- 1ul 2mM dNTPs * 8.8 = 8.8ul
- 1ul 25mM MgCl2 * 8.8 =8.8ul
- 0.25ul 10uM 1490 primer * 8.8 = 2.2ul
- 0.25ul 10uM 2198 primer * 8.8 = 2.2ul
- 0.1ul NEB Taq * 8.8ul = 0.88ul
- 4.4ul molecular grade water * 8.8 = 38.72ul
- Master mix was vortexed, spun down, and kept on ice
- 8ul of master mix was added to each tube of 8 strip tubes (kept on ice)
- 2ul of DNA from samples was added to their respective reaction tube, 2ul of molecular grade water was added to the negative control, and 1ul of water and 1ul of DNA was added to the positive control. (see table below)
| Tube # | Sample | ul DNA |
|---|---|---|
| 1 | 3mL-150 | 2 |
| 2 | 3mL-UP | 2 |
| 3 | 3mL-LOW | 2 |
| 4 | 3mL-WELL | 2 |
| 5 | Gel-Control | 2 |
| 6 | PFG-Ladder | 2 |
| 7 | Negative control | 2ul molecular grade water |
| 8 | Positive control | 1ul molecular grade water, 1ul DNA |
- Strip tubes were vortexed and spun down until no bubbles
- Tubes were placed in the PCR machine, in a new program COI, which is based off the one in Tom’s folder but with 30 cycles
- 94 degrees C 2 min
- 94 degrees C 20 sec
- 52 degrees C 20 sec
- 72 degrees C 1 min 30 sec
- 72 degrees C 5 minutes
- 12 degrees C hold
- bold fields are cycled 30 times
- Run time was 1 hour and 45 minutes
- Tubes were put in the 4 degree fridge overnight until gelling, tubes are labeled with primer name and date
20211117 p47 PCR
- Used the same samples, same order, same style mastermix for the p47 PCR
- The positive control is a from Kent and is a positive control for p47 (virus)
- All samples were kept on ice, along with primers, and enzymes
- Made master mix on ice:
- 1ul 10X NEB taq buffer * 8.8 = 8.8ul
- 1ul 2mM dNTPs * 8.8 = 8.8ul
- 1ul 25mM MgCl2 * 8.8 =8.8ul
- 0.25ul 10uM p47_F primer * 8.8 = 2.2ul
- 0.25ul 10uM p47_R primer * 8.8 = 2.2ul
- 0.1ul NEB Taq * 8.8ul = 0.88ul
- 4.4ul molecular grade water * 8.8 = 38.72ul
- Master mix was vortexed, spun down, and kept on ice
- 8ul of master mix was added to each tube of 8 strip tubes (kept on ice)
- 2ul of DNA from samples was added to their respective reaction tube, 2ul of molecular grade water was added to the negative control, and 1ul of water and 1ul of DNA was added to the positive control. (see table below)
| Tube # | Sample | ul DNA |
|---|---|---|
| 1 | 3mL-150 | 2 |
| 2 | 3mL-UP | 2 |
| 3 | 3mL-LOW | 2 |
| 4 | 3mL-WELL | 2 |
| 5 | Gel-Control | 2 |
| 6 | PFG-Ladder | 2 |
| 7 | Negative control | 2ul molecular grade water |
| 8 | Positive control | 1ul molecular grade water, 1ul DNA |
- Strip tubes were vortexed and spun down until no bubbles
- Tubes were placed in the PCR machine p47 program in the Maggie folder:
- 94 degrees C 5 minutes
- 94 degrees C 45 seconds
- 55 degrees C 1 minute
- 68 degrees C 1.5 minutes
- 68 degrees C 5 minutes
- Hold at 12 degrees C
- bold text is cycled through 30 times
- Run time was 2 hours
- Tubes were put in the 4 degree fridge overnight until gelling, tubes are labeled with primer name and date
20211202 Gel of PCR Products
- Microwaved pre-made gel
- Used the gel tray that has the green stripes and fist ~22 samples, I overpoured it a little so I made sure to stain it for a long time
- Loaded a 1kb plus ladder in the first well and in the well between the COI and the p47 samples, and between the 3mL-150 gel extraction sample
- Loaded all the COI samples, then the p47 samples
- Ran the gel for 40 min at 90V
- Put the gel in the EtBr stain for 1 hour and 15 minutes
- No amplification of either COI or p47
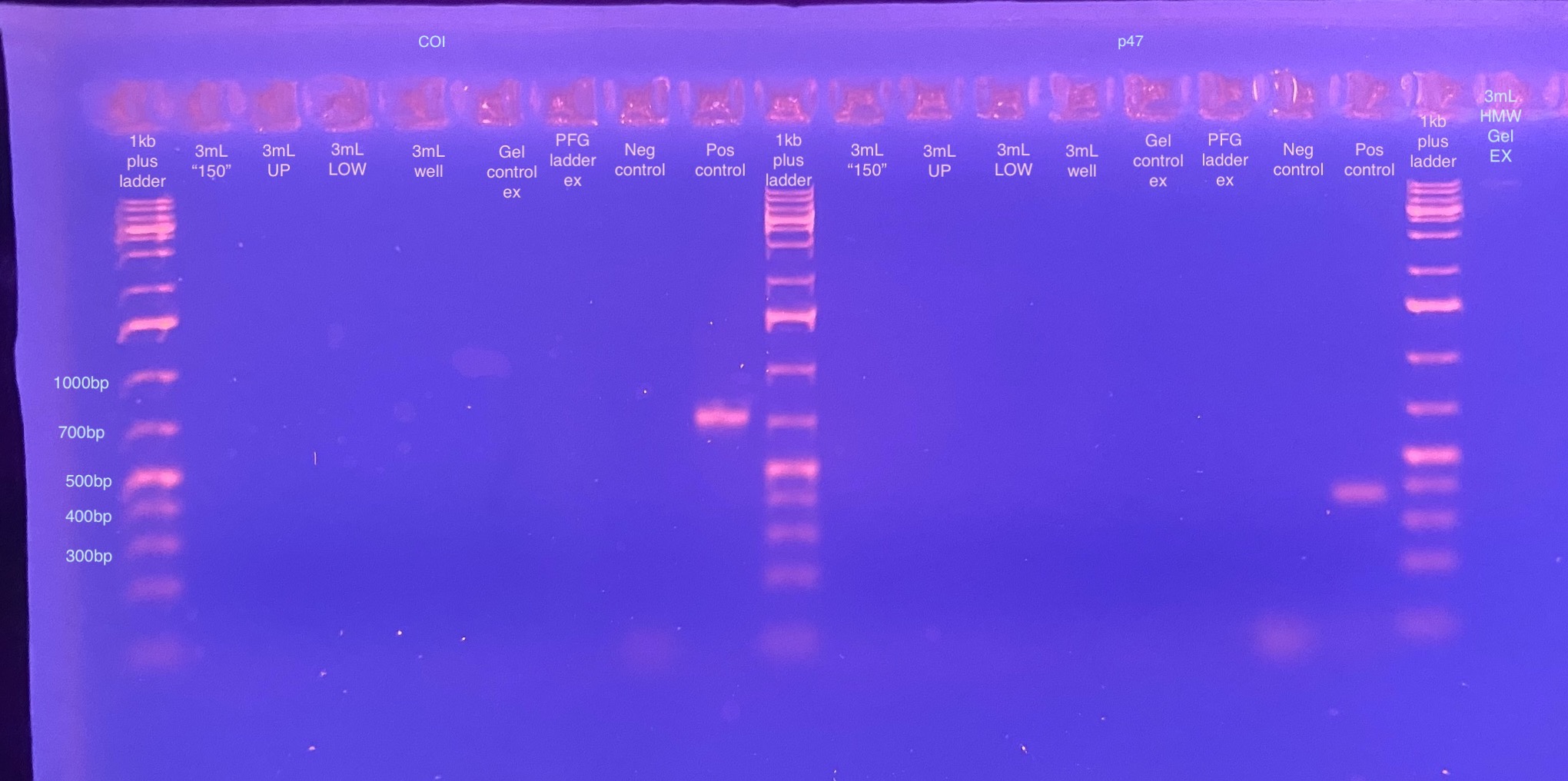
This could be for a few reasons. 1 is that COI is part of the mitochondria, it wouldn’t be in with HMW chunk of the extraction! So bad control gene. It could be possible that the DNA is fine and there’s just no virus in there. Another reason could be that the gel extraction samples have too much ethanol in them and the PCRs aren’t working at all, and there is virus in there. Or there could be not enough DNA in the PCRs to amplify (I put in less than 10ng for all of them). I should try these samples again with RPL 11 primers (what Kent uses for D. virilis amplification), as well as trying the original HMW extraction for both RPL 11 and p47. That will let me know if I should proceed or not with trying an overnight gel again.
Also the 3mL-150 gel extraction sample barely showed up (most had floated out of the well, likely because of ethanol contamination left over from the gel extraction kit), but it doesn’t look sheared? Or at least it’s 20kb in size or larger.