Troubleshooting DNA/RNA Extraction of Pentagona and Hystera Sea Stars 5
DNA/RNA Extractions on 3 Sea Star Tissue Samples Stored in DNA/RNA Shield, 3 Pentagona and 1 Hystera, 2 Samples With Dremel Homogenization, 2 With Tissuelyser
Notes
- Using the Zymo DNA/RNA Miniprep Plus kit
- Used Dremel with drill bit head for 20 seconds at lowest power setting (5) on the first two samples
- Used 1.4mm ceramic beads and the tissuelyser for 2 minutes at 25Hz for the second two samples
- No incubation, no proteinase K
Sample Preparation
- Samples:
- CPAB2-001 dremel
- CHSY-005 dremel
- CPAB-006 tissue lyser
- CPBP-002 tissue lyser
- Thawed samples on ice bucket
- Prepared forceps, foil, and razor blades
- Cleaned forceps with 10% bleach, DI water, 70% ethanol, and RNaseZap before and between each sample
- Cleaned dremel bit by bathing in beach, DI and ethanol, RNaseZap and touching only with forceps between each sample
- Made 1 1.5mL tube per sample with 300ul of DNA/RNA shield for each dremel sample
- Made 1 ceramic bead tube with 500ul DNA/RNA shield for each tissue lyser sample
- Cut a small piece of tissue for each sample then minced with the razor blade until it was flat and could come apart a little bit and placed in their respective homogenization tube
- CPAB2-001 tissue piece and minced piece

- CHSY-005 tissue piece and minced piece

- CPAB-006 tissue piece and minced piece

- CPBP-002 tissue piece and minced piece


- Dremel homogenized the first two samples (cleaning bit in between) for 20 seconds on 5 speed
- Before homogenization with the dremel:
- After homogenization with the dremel:
- Put the bead tubes in the TissueLyser for 2 minutes at 25Hz (balenced with empty bead tubes)
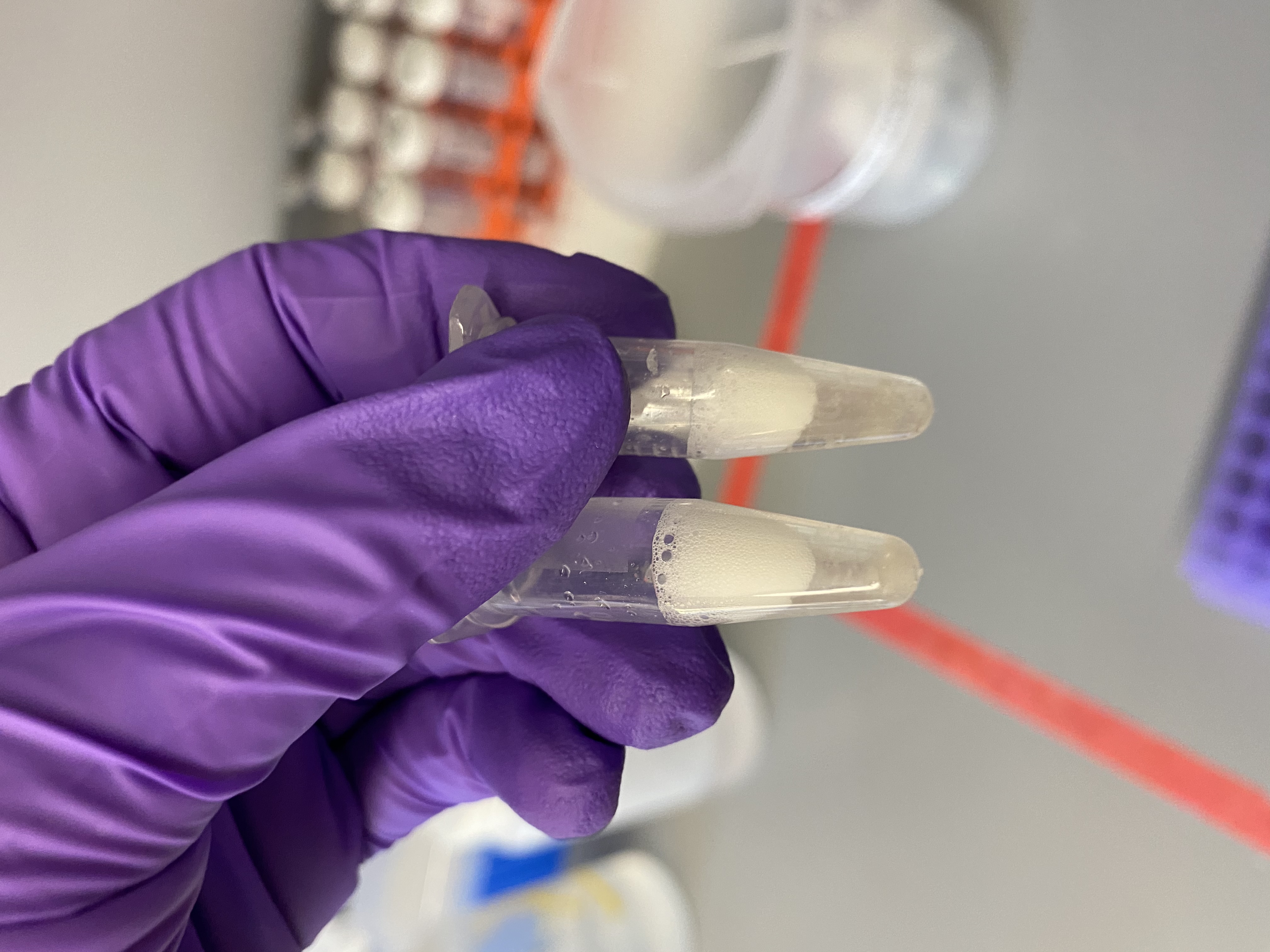
- After TissueLyser homogenization:
- Spun down the bead tubes in the minifuge to remove bubbles
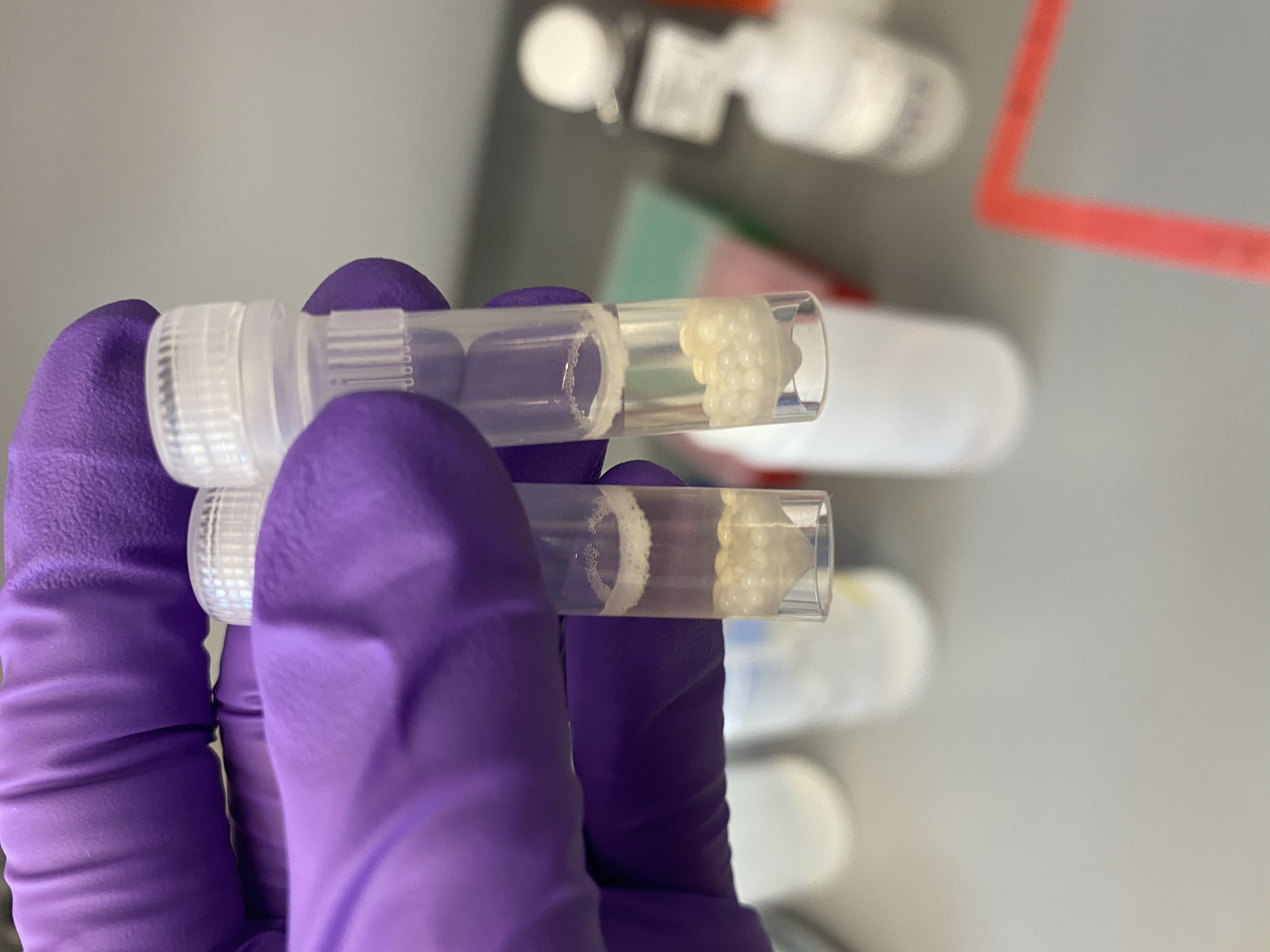
- Centrifuged the dremel-homogenized tubes in the centrifuge at max speed for 2 min to pellet debris
- Still some floating debris in the dremel-tubes after centrifugation, like the previous dremel extraction:
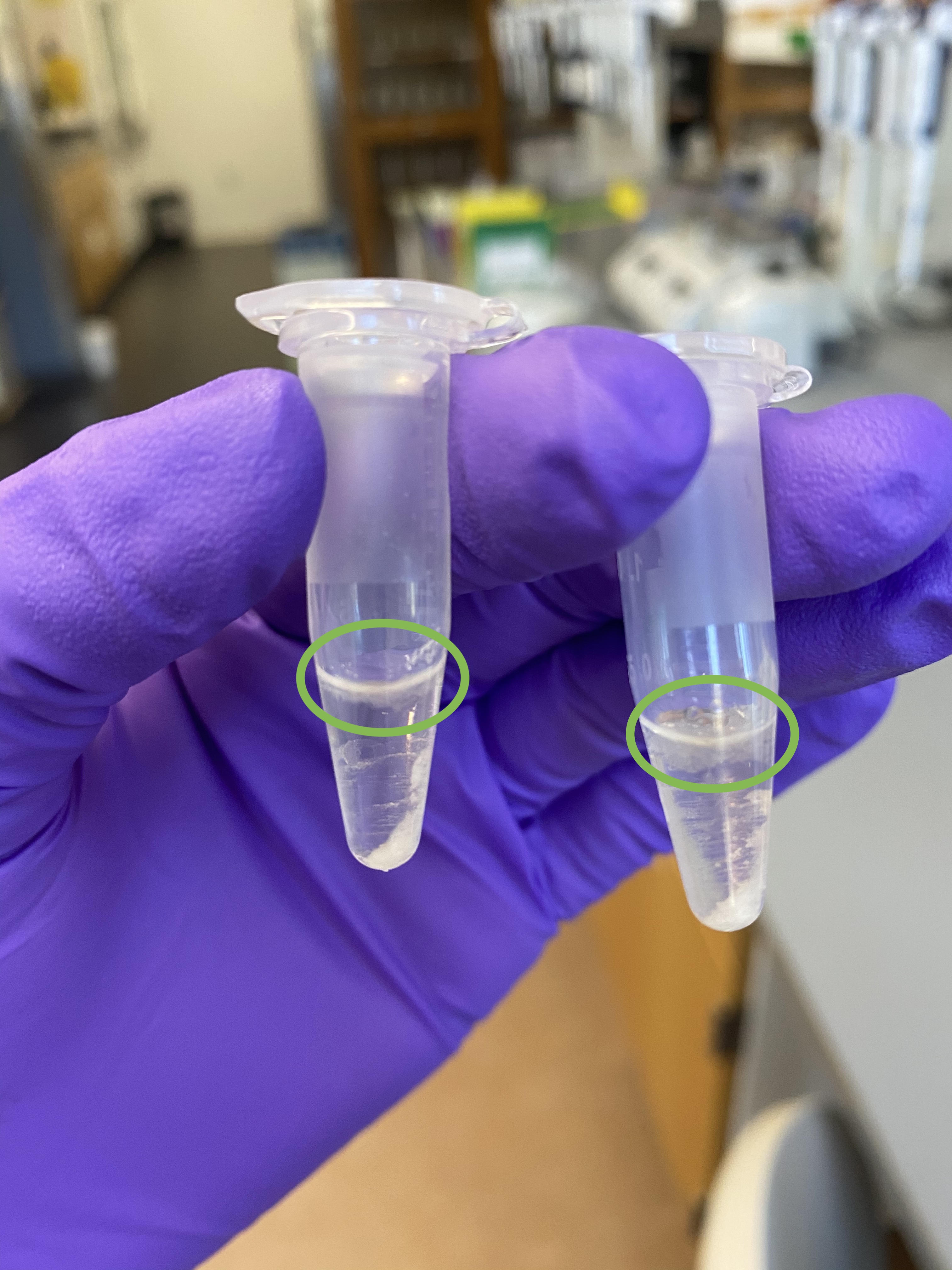
- Avoided all debris and pipetted supernatant from each tube into each a new 1.5mL tube
- Added equal volume DNA/RNA lysis buffer to each tube (300ul for dremel tubes, 450ul for tissue lyser tubes)
DNA Extraction
- Flicked and inverted tubes to mix and spun down
- Warmed 10mM Tris HCl pH 8 and nuclease free water in the thermomixer at 70 degrees C
- Added 700ul of the liquid to yellow spin columns and collection tubes
- Centrifuged 16,000rcf for 30 seconds
- Saved the flow through in new 5mL tubes
- Added the remaining liquid to the yellow spin columns
- Centrifuged 16,000rcf for 30 seconds
- Saved the flow through in the 5mL tubes
- Added 400ul DNA/RNA prep buffer to the yellow spin columns
- Centrifuged 16,000rcf for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 700ul DNA/RNA wash buffer to the yellow spin columns
- Centrifuged 16,000rcf for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 400ul DNA/RNA wash buffer to the yellow spin columns
- Centrifuged 16,000rcf for 2 minutes
- Transferred spin columns to new 1.5mL tubes for each sample labeled as final DNA tubes
- Discarded flow through and collection tubes
- Added 50ul warmed 10mM Tris HCl to the spin columns gently by dripping over the filter
- Incubated at room temp for 5 minutes
- Centrifuged 16,000rcf for 30 seconds
- Added another 50ul warmed 10mM Tris HCl to the spin columns gently by dripping over the filter
- Incubated at room temp for 5 minutes
- Centrifuged 16,000rcf for 30 seconds
- Discarded the spin columns
- Made strip tubes with 7ul of each sample in them for QC
- Kept tubes on ice bucket then stored in -20 degree freezer
RNA Extraction
- Added equal volume (600ul and 750ul) 100% ethanol to each 5mL tube
- Vortexed and spun down
- Added 700ul of the liquid to green spin columns and collection tubes
- Centrifuged 16,000rcf for 30 seconds
- Discarded flow through (Zymo kit waste)
- Repeated addition for the remaining liquid (2 times) to the green spin columns
- Centrifuged 16,000rcf for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 400ul DNA/RNA wash buffer to the green spin columns
- Centrifuged 16,000rcf for 30 seconds
- Discarded flow through (Zymo kit waste)
- Created the DNase mix:
- 75ul DNA digestion buffer * 4 = 300ul
- 5ul DNase I * 4 = 20ul
- Flicked and spun down mix
- Added 80ul DNAse mix to each green spin column filter
- Incubated for 15 minutes at room temp
- Added 400ul DNA/RNA prep buffer to the green spin columns
- Centrifuged 16,000rcf for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 700ul DNA/RNA wash buffer to the green spin columns
- Centrifuged 16,000rcf for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 400ul DNA/RNA wash buffer to the green spin columns
- Centrifuged 16,000rcf for 2 minutes
- Transferred spin columns to new 1.5mL tubes for each sample labeled as final RNA tubes
- Discarded flow through and collection tubes
- Added 50ul warmed nuclease free water to the spin columns gently by dripping over the filter
- Incubated at room temp for 5 minutes
- Centrifuged 16,000rcf for 30 seconds
- Added another 50ul warmed nuclease free water to the spin columns gently by dripping over the filter
- Incubated at room temp for 5 minutes
- Centrifuged 16,000rcf for 30 seconds
- Discarded the spin columns
- Made strip tubes with 5ul of each sample in them for QC
- Kept tubes on ice bucket then stored in -80 degree freezer
QC
Broad Range Qubit for DNA and RNA
- Followed Qubit protocol
- DNA
| Sample | Reading 1 (ng/ul) | Reading 2 (ng/ul) | Average DNA (ng/ul) |
|---|---|---|---|
| Standard 1 | 187 RFU | - | - |
| Standard 2 | 21138 RFU | - | - |
| CPAB2-001 | 13.7 | 13.5 | 13.6 |
| CHSY-005 | 20.8 | 20.4 | 20.6 |
| CPAB-006 | 25.6 | 25.4 | 25.5 |
| CPBP-002 | 32.8 | 32.6 | 32.7 |
- RNA
| Sample | Reading 1 (ng/ul) | Reading 2 (ng/ul) | Average RNA (ng/ul) |
|---|---|---|---|
| Standard 1 | 398 RFU | - | - |
| Standard 2 | 8374 RFU | - | - |
| CPAB2-001 | too low | - | - |
| CHSY-005 | too low | - | - |
| CPAB-006 | 14.6 | 14.4 | 14.5 |
| CPBP-002 | 15.2 | 14.8 | 15 |
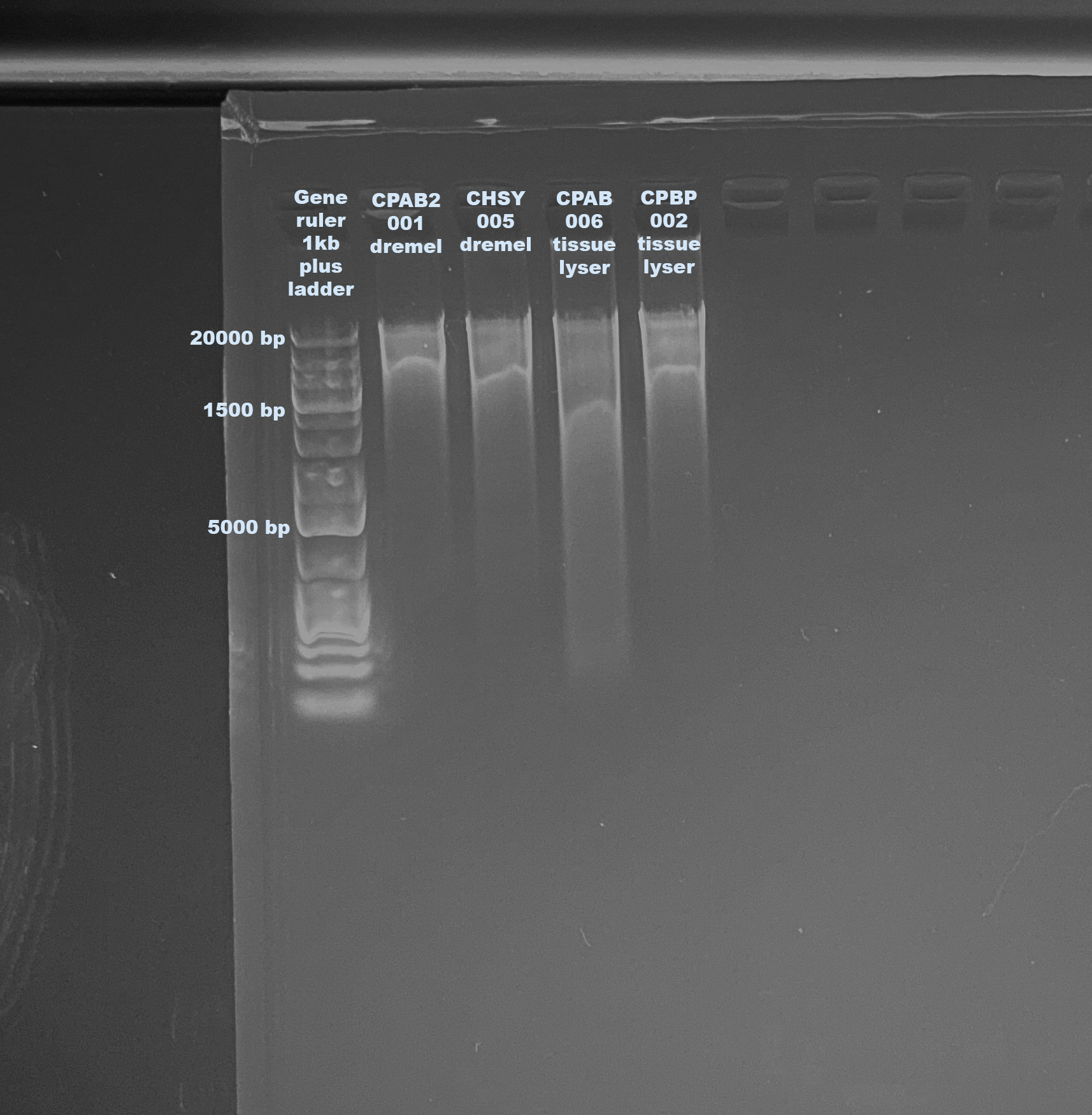
1% Agarose Gel for DNA Quality
- Followed gel protocol
RNA TapeStation
Notes
RNA quality is really good, just yield is low. DNA quality looks similar between the two methods.
Written on November 5, 2020