Test RNA Extraction from Sea Stars with Zymo Direct-zol Kit
Testing RNA Extraction and Yield from Sea Stars with the Zymo Direct-zol RNA Miniprep Plus Kit
Notes:
- All steps of the extraction other than the Tissuelyser and the centrifugation were done in the hood
- All tips used were filter tips
- Gloves, lab coat, mask, and glasses were worn at all times
- Spaces and plastic materials were wiped with 10% bleach and RNaseZap before use
- Forceps were cleaned with 10% bleach, DI water, 70% Ethanol and RNaseZap between uses
- Samples were cut on foil
Sample Prep
- Samples thawed and temporary tube numbers:
- CPAB002 -> # 3
- CHSB005 -> # 4
- Took out sample 1 and placed small chunk in an empty bead tube
- Immediately added 800ul of tri-reagent to the bead tube (# 2), following recommendations from the Zymo protocol for solid tissues
- Took out sample 2 and placed small chunk in another empty bead tube

- Immediately added 800ul of tri-reagent to the second bead tube (# 3)
- I noticed that the liquid (DNA/RNA Shield) in the first tube had a lot of tissue chunks in it and I wondered if the tissue was mostly in the liquid and not in the big piece (seemed mostly like ossicles) so I added 400ul of the liquid to another bead tube (# 5)
- Sample tube # 2 had completely clear liquid but I also made a bead tube with 400ul of it just to be consistent (# 6)

- Added 400ul of tri-reagent to each of the bead tubes with liquid, following the recommendations from the Zymo protocol for liquid samples (1:1 ratio)
- Balanced the Tissuelyser with 4 tubes in each side (some empty) and ran it at 30Hz for 2 minutes (combination of recommendations from the Zymo protocol: “high speed” for 30-60 seconds, and the tissuelyer RNA from tissue recommendation 4 minutes at 30Hz)
- Briefly spun down tubes in the minifuge to collect beads at the bottom
- Tubes 3, 5, and 6 were opaque at this point, and the Zymo protocol says opaque liquid means incomplete lysis, and to add more tri-reagent and homogenize again
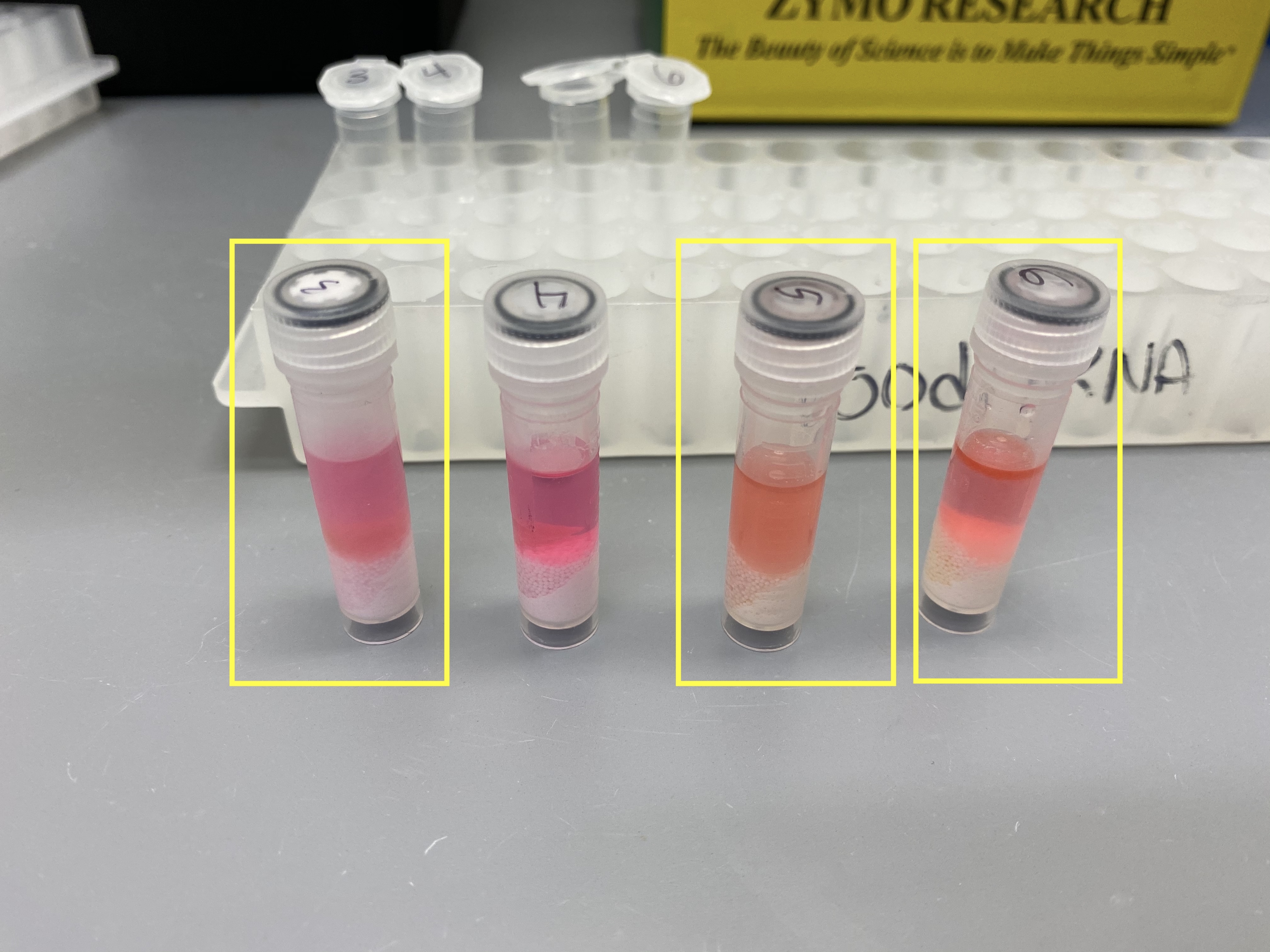
- So I added 400ul more tri-reagent to those 3 tubes and used the tissuelyser again for 2 minutes at 30Hz
- Briefly spun down the tubes again and now only # 3 looked opaque. There wasn’t enough room in the tube for more tri-reagent so I transferred 300ul of the liquid in tube # 3 to a new 1.5mL tube (3B) and added what was left of the tri-reagent (300ul) each to tube # 3 and tube # 3B and vortexed those two tubes and spun them down
- Then I transferred all the liquid (carefully not sucking up beads) from each tube (3, 4, 5, 6 and 3B) into new 5mL tubes, measuring the volume of liquid
- 3: 850ul
- 4: 600ul
- 5: 900ul
- 6: 800ul
- 3B: 700ul
- Added equal volume 100% ethanol to each 5mL tube and invert to mix
- Used the large centrifuge to spin down the tubes briefly, then I noticed that in all the tubes there was a white pellet at the bottom, probably the really fine beads! I centrifuged them at max speed for a minute to pellet any beads. Now all the liquid was clear in all of the tubes!
Tube 3 after 1st spin, still foggy but a lot of beads present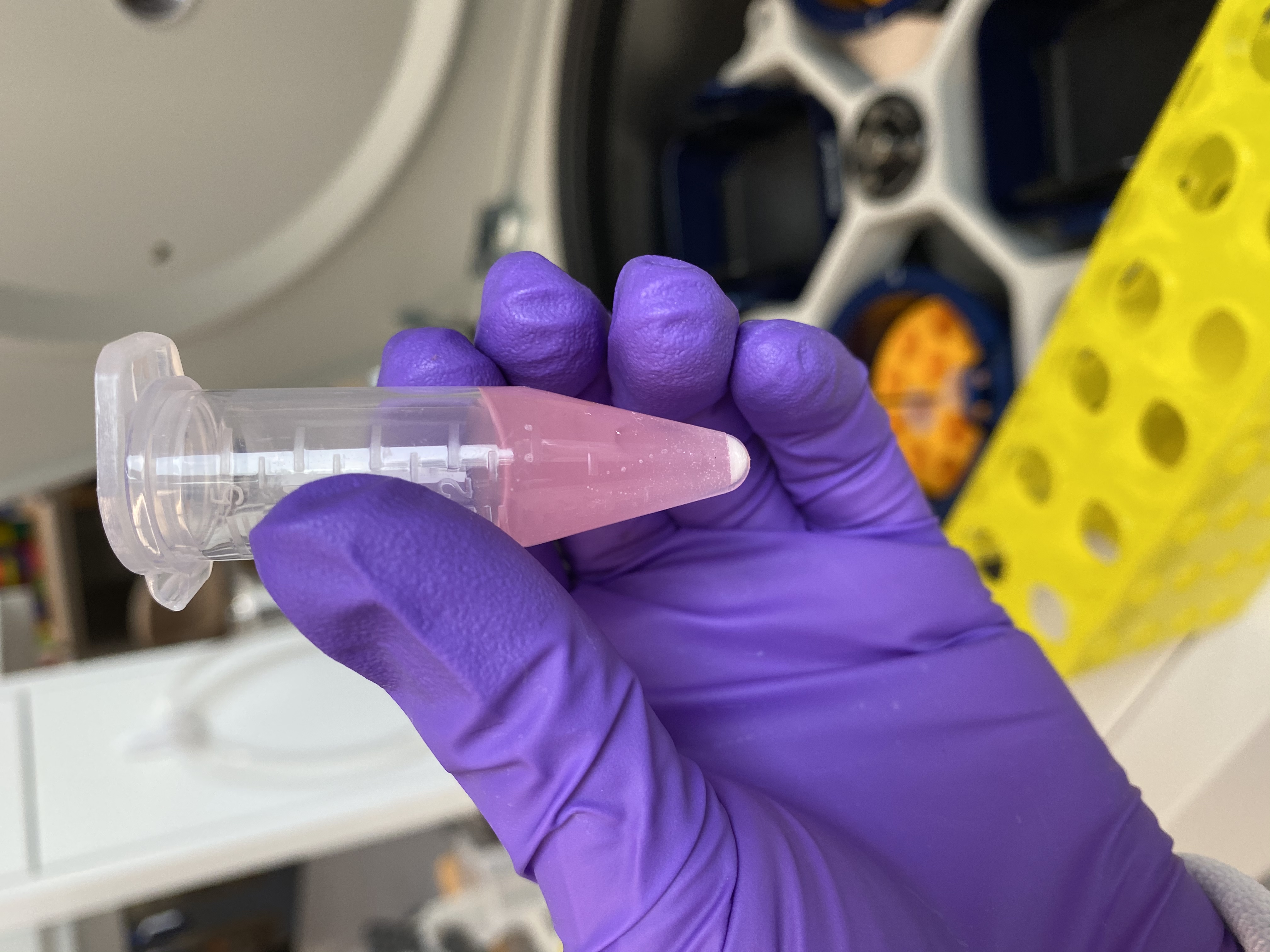
Even already clear tube 6 had beads
Extraction
- Without sucking the beads up at the bottom I added 700ul from each 5mL tube to their respective spin columns and centrifuged at 16,000rcf for 30 seconds and discarded the flowthrough in the tri-reagent waste after
- I repeated the spins until all the liquid from the samples had gone through their columns
- I added 400ul of RNA wash buffer to each column, centrifuged, and discarded the flow through in the proper waste
- Created the DNase I mix: 375ul DNA digestion buffer and 25ul DNase 1
- Added 80ul of the DNase I mix to each filter by dripping
- Incubated for 15 minutes at room temp
- Added 400ul of direct-zol pre-wash to each column, centrifuged, and discarded the flowthrough in the proper waste
- Repeated the last step
- Added 700ul of the RNA wash buffer to each column, centrifuged for 1 minute, and discarded the flow through in the proper waste
- Transferred columns to new labeled 1.5mL tubes
- Added 50ul of RNase-free water to each column by dripping and centrifuged for 30 seconds
- Repeated the last step
- Aliquoted 5ul for qubit and gel into strip tubes and stored the final tubes in the grey -80
Qubit
- Followed the Qubit protocol for broad range RNA
- Used the 0ng/ul and 100ng/ul standards
| standard 1 | standard 2 | 3 | 4 | 5 | 6 | 3B |
|---|---|---|---|---|---|---|
| 387 RFU | 8548 RFU | 27.4ng/μl | 10.6ng/μl | 15.5ng/μl | too low | 17.2ng/ul |
TapeStation
- Followed the RNA TapeStation protocl
- I did not run tube # 6
Written on September 15, 2020