RNA Extraction and cDNA Library Prep for Pentagona and Hystera Sea Stars
Extraction RNA and Stranded mRNA Library Prep for Pentagona and Hystera Sea Stars for Use in Genome Building
RNA Extraction
Used Zymo Research Quick DNA/RNA Miniprep Plus Kit for extraction (probably should just have used the RNA one) and the Zymo bashing beads for homogenization. Roughly followed Emma Strand’s Soft Homogenization Protocol for ideas on how to homogenize samples because I’d never worked with sea stars before and Emma consistently gets good RNA with her protocol. Additionally I looked at this version of Mo’orea coral extractions which was the only time when I got good RNA out of those samples, and they are also tissue preserved in DNA/RNA shield, so that is why I had no incubation step.
- Took out 1 P6 and 1 H6 sample (tissue in DNA/RNA shield) and thawed on ice
- Filled two ZR bashing bead tubes with 700μl DNA/RNA Shield and named the tubes PA and HA
- Sterilized foil, scalpel, foil, and forceps. Took sample out of tube and cut in half on foil. Placed one half in the new bashing bead tube
- Vortexed each sample for 1 minute max speed
- Let sit after votexing: very bubbly. HA liquid was a lot more orange than PA. There was still a pretty prominent ossicle chunk left in there
- Aspirated all liquid I could get off into new labeled 1.5mL tubes
- Took 300μl of that into new 1.5mL tubes going forward
- Added 30μl prok digestion buffer
- Added 15μl Pro K
-
Vortexed and spun down tubes
- Equal volumes of DNA/RNA Lysis Buffer were added to each sample tube: 345μl
- Mixed samples by flicking and spinning down
- ~700µl of sample was gently added to Yellow DNA spin columns
- Centrifuged columns at 16000 rcf for 30 seconds
- Flow-through was transferred to new 1.5 mL tubes for RNA
- Added 400µl DNA/RNA Prep Buffer gently to the yellow DNA spin columns
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 700µl DNA/RNA Wash Buffer gently to the yellow DNA spin columns
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 400µl DNA/RNA Wash Buffer genetly to the yellow DNA spin columns
- Centrifuged at 16,000 rcf (g) for 2 minutes
- Discarded flow through (Zymo kit waste)
- Transferred yellow columns to new 1.5mL microcentrifuge tubes
- Add 50µl warmed 10mM Tris HCl to each yellow DNA column
- Incubated at room temp for 5 minutes
- Centrifuged at 16,000 rcf (g) for 30 seconds
-
Repeated last three steps for a final elution volume of 100µl
- Added equal volume 100% EtOH to the 1.5mL tubes labeled for RNA containing the original yellow column flow through: ~700µl
- Vortexed and spin down to mix
- Added 700µl of that liquid to the green RNA spin columns
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 700µl to the green RNA spin columns (the rest from the 1.5mL RNA tubes)
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 400µl DNA/RNA Wash Buffer gently to each green RNA column
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Made DNase I treatment master mix:
- 75µl DNA Digestion buffer x 2 = 150µl
- 5µl DNase I x 2 = 10µl
- Added 80µl DNase I treatment master mix directly to the filter of the green RNA columns
- Incubated at room temp for 15 minutes
- Added 400µl DNA/RNA Prep Buffer gently to each column
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 700µl DNA/RNA Wash Buffer gently to the yellow DNA spin columns
- Centrifuged at 16,000 rcf (g) for 30 seconds
- Discarded flow through (Zymo kit waste)
- Added 400µl DNA/RNA Wash Buffer genetly to the yellow DNA spin columns
- Centrifuged at 16,000 rcf (g) for 2 minutes
- Discarded flow through (Zymo kit waste)
- Transferred green columns to new 1.5mL microcentrifuge tubes
- Added 50µl warmed DNase/RNase free water to each green RNA column by dripping slowly directly on the filer
- Incubated at room temp for 5 minutes
- Centrifuged at 16,000 rcf (g) for 30 seconds
- ed last three steps for a final elution volume of 100µl
- Labeled 1.5mL tubes on ice afterwards, and aliquoted 5µl into PCR strip tubes to save for Qubit and Tape Station to avoid freeze-thaw
- Stored all tubes in the -80
Qubit and TapeStation
| Sample | DNA Standard 1 (RFU) | DNA Standard 2 (RFU) | DNA 1 (ng/µl) | DNA 2 (ng/µl) | Average DNA | RNA Standard 1 (RFU) | RNA Standard 2 (RFU) | RNA 1 (ng/µl) | RNA 2 (ng/ul) | Average RNA |
|---|---|---|---|---|---|---|---|---|---|---|
| PA | 181 | 20099 | 66 | 65.4 | 65.8 | 385 | 10633 | 18.8 | 18.4 | 18.6 |
| HA | 181 | 20099 | 16.9 | 16.7 | 16.8 | 385 | 10633 | 19.6 | 19.2 | 19.4 |
Full TapeStationResults:
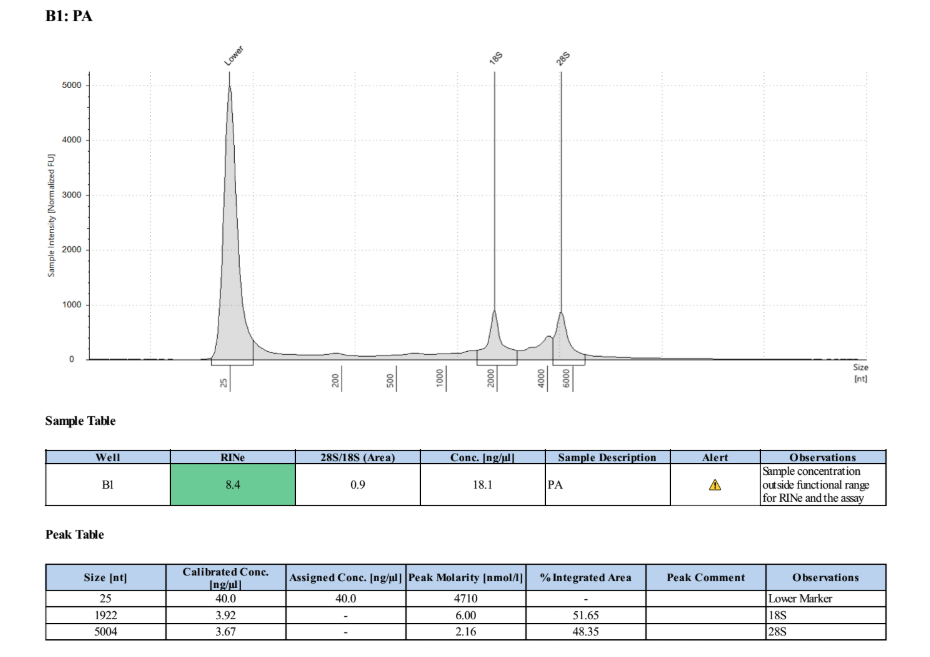
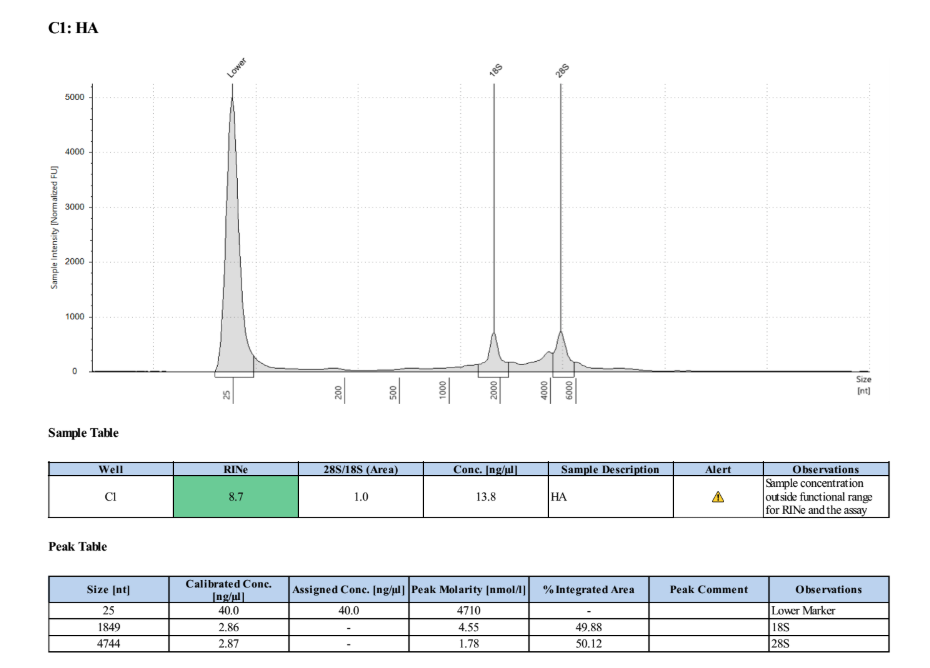
Stranded mRNA Seq Prep
Samples are concentrated enough for the library prep, so they were vacufuged for ~2 hours and then re-qubited. PA: 42.8ng/µl. HA: 47.4ng/µl.
Capture
- Took out mRNA capture beads, mRNA bead binding buffer, and mRNA bead wash buffer from 4 degree and let get to room temp
- Resuspended mRNA capture beads by pipetting up and down
- Calculated the number of beads needed:
26.25μl beads * 2 samples = 52.5μl - Pipetted 52.5μl mRNA capture beads into a new PCR tube and put on the plate magnet stand
- Removed supernatant
- Added 52.5μl of bead binding buffer and pipetted to mix off magnet
- Put back on magnet and removed as much supernatant as possible
- Added 52.5μl of bead binding buffer again and pipetted to mix off magnet
- Put back on magnet and again removed as much supernatant as possible
- Added 52.5μl of bead binding buffer and pipetted to mix off magnet
- Added 1μg of RNA from each sample into new PCR tubes and filled the rest up to 25μl with nuclease free water:
- PA: 23.36μl RNA and 1.64μl nuclease-free water
- HA: 21.28μl RNA and 3.72μl nuclase-free water
- Added 25μl of the resuspended mRNA capture beads to each sample tube
- Placed tubes in the 1st mRNA capture program in the thermocycler
- Placed tubes on the magnet plate and removed all supernatant when the solution went clear
- Removed tubes from the magnet plate and resuspended beads in 100μl of bead wash buffer
- Placed tubes on the magnet plate and removed all of the supernatant when the solution went clear
- Resuspended beads in 25μl of RNase-free water off magnet
- Placed tubes in the 2nd mRNA capture program in the thermocycler
- Took tubes out of the thermocycler and added 25μl of bead binding buffer to each tube and pipetted to mix
- Incubated the tubes on the shaker at 200rpm for 5 minutes
- Made 1X Fragment, Prime, and Elute Buffer on ice bucket:
5.5μl water * 2.2 = 12.1μl
5.5μl 2X FPE buffer * 2.2 = 12.1μl - Placed tubes on the magnet plate and removed supernatant when the solution went clear
- Resuspended beads off magnet in 11μl 1X FPE buffer
- Placed tubes in the 4 degree fridge overnight (safe stopping point)
Fragmentation
- Took tubes out of fridge and put in thermocycler for RNA fragmentation program 300-400bp (6 minutes at 85 degrees)
- Made 1st Strand Synthesis Master Mix on ice:
5.5μl 1st strand synthesis buffer * 2.2 = 12.1μl
.5μl KAPA script * 2.2 = 1.1μl - IMMEDIATELY placed tubes on magnet plate once the program was finished (some tubes had popped open)
- Removed 10μl of clear supernatant and placed in new PCR strip tubes on ice
1st Strand Synthesis
- Added 5μl of the 1st strand synthesis master mix to each tube and pipetted to mix
- Placed in thermocycler 1st strand synthesis program
- Made 2nd Strand Synthesis and Marking Master Mix on ice:
15.5μl 2nd strand marking buffer * 2.2 = 34.1μl
1μl second strand enzyme * 2.2 = 2.2μl - Removed tubes from the thermocycler and placed on ice
2nd Strand Synthesis
- Added 15μl of the 2nd strand synthesis and marking master mix and pipetted to mix
- Put in thermocycler 2nd strand synthesis program
- Took KAPA Pure Beads out of the 4 degree and swirled them to mix and let get to room temp
- Took tubes out of the thermocycler and added 54μl KAPA Pure Beads, pipetting to mix
- Incubated tubes on shaker for 15 minutes
- Made A-tailing Immediately Master Mix on ice:
12μl water * 2.2 = 26.4μl
1.5μl 10X A-tailing buffer * 2.2 = 3.3μl1.5μl A-Tailing enzyme * 2.2 = 3.3μl - Made fresh 80% EtOH for that day
- Placed tubes on magnet plate and removed 80μl clear supernatant
- Added 200μl 80% EtOH to each tube
- Removed EtOH from each tube
- Added 200μl 80% EtOH to each tube
- Removed ALL EtOH from each tube, using a p20 to get extras and droplets of EtOH
A-Tailing
- Waited ~30 seconds or less and resuspended the beads in 15μl of the A-tailing Immediately Master Mix
- Put tubes in the thermocycler A-tailing program
Adapter Ligation
- Made the Adapter Ligation Master Mix:
8μl nuclease-free water * 2.2 = 17.4μl
7μl ligation buffer * 2.2 = 15.4μl
2.5μl DNA ligase * 2.2 = 5.5μl - Added 17.5μl of the ligation master mix to each tube out of the theromocylcer
- Added 2.5μl of the diluted/annealed/working stock 700nM No-Barcode (NOBO) adapters to each sample
- Pipetted to mix
- Placed tubes on shaker for ~1hour room temp
2 Cleanups
- Added 35μl of room temperature PEG to each sample and pipetted to mix
- Placed on shaker for 15 minutes
- Made fresh 80% EtOH
- Placed tubes on magnet plate
- Removed 67μl of supernatant from each tube
- Added 100μl 80% EtOH to each tube
- Removed 100μl of supernatant from each tube
- Added 100μl 80% EtOH to each tube
- Removed ALL of the supernatant from each tube, using a p20 pipette tip to get rid of droplets
- Resuspended beads in 25μl of 10mM Tris HCl pH 8
- Placed tubes on shaker for 5 minutes
- Added 25μl of room temperature PEG to each tube and pipetted to mix
- Placed on shaker for 15 minutes
- Made fresh 80% EtOH
- Placed tubes on magnet plate
- Removed 45μl of supernatant from each tube
- Added 100μl 80% EtOH to each tube
- Removed 100μl of supernatant from each tube
- Added 100μl 80% EtOH to each tube
- Removed ALL of the supernatant from each tube, using a p20 pipette tip to get rid of droplets
- Resuspended beads in 11μl of 10mM Tris HCl pH 8
- Placed tubes on shaker for 5 minutes
- Placed tubes on magnet stand and removed 10μl of clear supernatant to new tubes
Library Amplification
To increase diversity of index sequences in sequencing (more color variation in the machine) the libraries were split at this point to get two different index combos per sample: PAmp1, PAmp2, HAmp1, and HAmp2. This means only 5μl of the above sample went in to each PCR, so the extra volume to 25μl was made up with water and the primer volume added was halved to keep the ratio the same.
- Made Library Amplification Master Mix:
12.5μl KAPA HotStart Ready Mix x 4.2 = 52.5μl
6μl nuclease-free water x 4.2 = 25.2μl - Made PCR reaction tubes with these reagents/samples:
| Sample | Amount of cDNA | Library MM | index 1 | index 2 |
|---|---|---|---|---|
| PAmp1 | 5μl PA | 18.5μl | .75μl 505 | .75μl 709 |
| PAmp2 | 5μl PA | 18.5μl | .75μl 502 | .75μl 710 |
| HAmp1 | 5μl HA | 18.5μl | .75μl 503 | .75μl 705 |
| HAmp2 | 5μl HA | 18.5μl | .75μl 511 | .75μl 703 |
- Pipetted to mix and placed in thermocyler 12 cycle PCR program (normal for this prep)
- Took out KAPA beads
- Did a 1X bead cleanup: 25μl beads to each sample
- Performed normal bead clean up (see above for examples)
- Resuspended and eluted beads in 22μl 10mM Tris HCl
Broad Range Qubit and TapeStation
PAmp1: 111ng/μl PAmp2: 102ng/μl HAmp1: 103ng/μl HAmp2: 101ng/μl
Full TapeStation Report
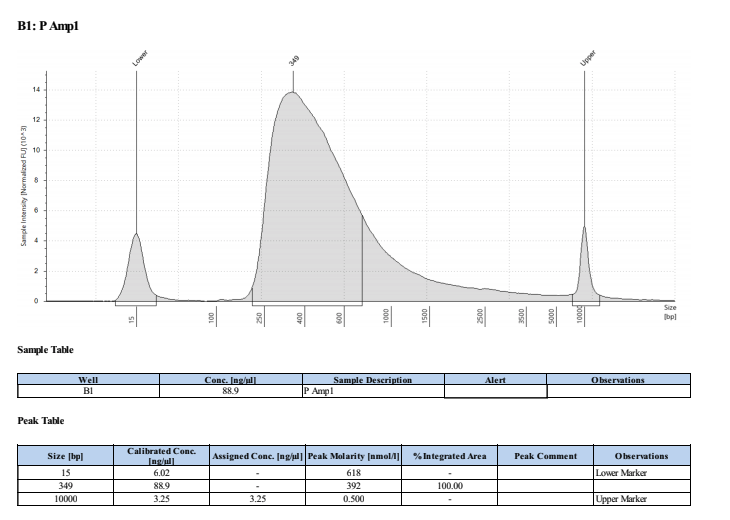
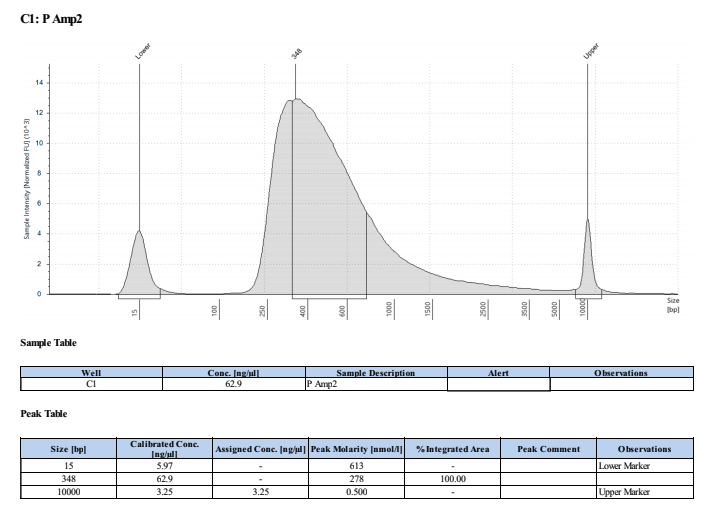
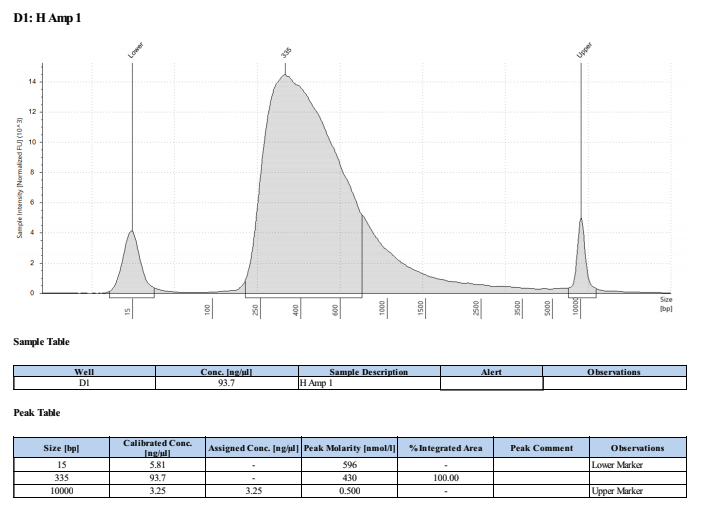
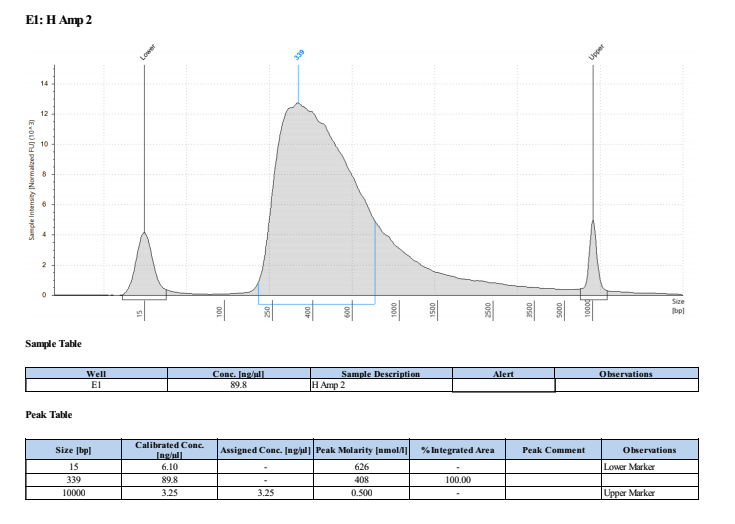
These worked well but they are smaller than we expected/wanted. Looks like they fragmented the RNA to ~200bp not 300bp…. Still sent to Seq tho!