Library Prep of pentagona HMW DNA for minION Sequencing
Using the Oxford Nanopore Ligation Sequencing Kit and the NEB Companion Module for Oxford Nanopore Technologies Ligation Sequencing on two HMW DNA pentagona samples
Two samples were chosen, because of struggles to get enough DNA after size selection, one sample was a pool of previous extractions post-BluePippin size selection: x x Samples PG1, PG2, PG3, PG1S, and PG2S were pooled for a combined library, and PG4 had enough DNA for a library prep of its own. Pooling samples was fine because all of the tissues and extractions were from the same individual.
Sample PG4 had a 0.45X bead cleanup after size selection, and all the samples pooled together went through a 1X bead cleanup and both were resuspended in 50µl nuclease-free water.
Some steps in this are modified from the original protocol. Modifications from J. Puritz.
DNA Repair and End-prep
- Thawed DCS (DNA control sequence) from the nanopore kit a room temp and once it was thawed it went on ice
- Thawed FFPE DNA Repair Buffer, FFPE DNA Repair Mix, Ultra II End-prep reaction buffer, and Ultra II End-prep enzyme mix on ice. From NEB kit
- Made master mix for DNA repair and end prep:
- 1µl DNA CS x 2.1 = 2.1µl
- 3.5µl FFPE DNA repair buffer x 2.1 = 7.35µl
- 2µl FFPE DNA repair mix x 2.1 = 4.2µl
- 3.5µl Ultra II end prep reaction buffer x 2.1 = 7.35µl
- 3µl Ultra II end prep enzyme mix x 2.1 = 6.3µl
- Pipetted to mix master mix
- Added 13µl of master mix to each sample tube in strip tubes (PG4 and PGComb)
- Flicked tubes to mix with DNA then spun down in minifuge
- Placed in thermocycler Nanopore Incubation program: 30min at 20 degrees C then 30min at 65 degrees C
Bead Cleanup
- Took out KAPA Pure beads, swirled to resuspend, and let come to room temp
- Made fresh 80% ethanol
- Took sample tubes out of the thermocyler and added 60µl beads to each tube
- Flicked tubes to mix then spun them down on a minifuge
- Placed tubes on shaker for 30 minutes at 300rpm
- Placed tubes on magnet plate and waited 5 minutes
- Removed and saved supernatant without disturbing the beads
- Gently added 200µl 80% EtOH
- Removed and discarded wash supernatant
- Added 200µl 80% EtOH
- Removed and discarded wash supernatant
- Spun down tubes with minifuge and placed back on magnet
- Removed any remaining supernatant
- Let beads dry maybe 30 seconds
- Resuspended pellet in 61µl nuclease-free water: flicked and spin down, flicked and spun down. No pipette mixing
- Incubated tubes room temp stagnant for 30 minutes
- Incubated tubes room temp shaking for at least 30 minutes
- Kept liquid with beads, placed on magnet plate and waited until clear
- Broad Range Qubit of samples:
- PGC: 24.1ng/µl
- PG4: 20.4ng/µl
Adapter Ligation
- Thawed AMX (Adapter Mix) and T4 DNA Ligase (NEB kit) on ice
- Thawed LNB (Ligation Buffer) at room temp, spun down, pipetted to mix, then placed on ice
- Thawed EB (Elution Buffer) at room temp, vortexed, spun down and put on ice
- Thawed LFB (Long Fragment Buffer) at room temp, vortexed, spun down and put on ice
- Made ligation and adapter master mix:
- 25µl LNB x 2.1µl = 52.5µl
- 10µl T4 DNA ligase x 2.1 = 21µl
- 5µl AMX x 2.1 = 10.5µl
- Flicked tube to mix (ligase!!) then spun down
- Added 40µl ligation and adapter mix to each tube with the DNA with the beads
- Flicked to mix the tubes then spun down
- Incubated tubes on shaker at room temp for 30min at 200rpm
Bead Cleanup
- Took out beads and let get to room temp
- After incubation, spun down tubes and added 40µl beads to each sample tube
- Flicked tubes to mix then spun down
- Incubated tubes on shaker at room temp for 30min at 300rpm
- Placed on magnet plate after incubation
- Waited 5 minutes
- Removed clear supernatant without disturbing beads and saved just in case
- Took tubes off magnet and added 250µl LFB
- Flicked tube to mix and spun down
- Placed back on magnet
- Removed and saved supernatant just in case
- Took tubes of magnet and added 250µl LFB
- Flicked tubes to mix and spin down
- Placed back on magnet
- Removed and saved supernatant
- Spun down tubes
- Removed any residual supernatant
- Removed tubes from magnet and added 15µl EB
- Flicked tubes to mix then spun down
- Placed tubes on shaker at 300rpm at room temp overnight
Quantify Libraries:
- PGC: 73.8ng/µl
- PG4: 58ng/µl
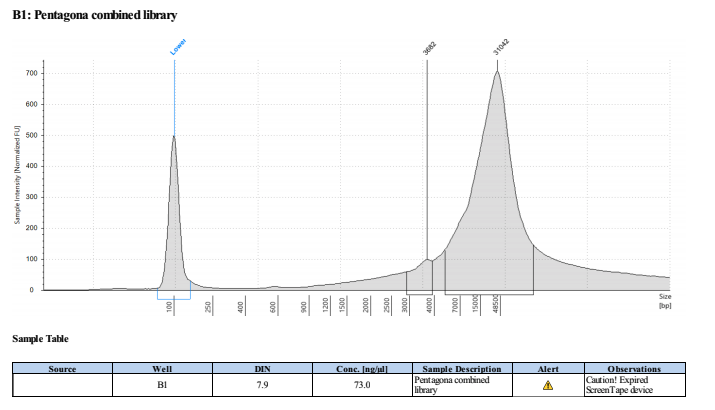
Libraries weren’t tapestationed before we put them on the minION, but the PGC library had a lower N50 than the PG4 library, so after I ran it on the tapestation. There are some 3kb fragments in there which there shouldn’t really have been…