Second Extraction Pentagona HMW Size Selection with BluePippin
Using the BluePippin High Pass Plus Cassette, >15 kb DNA size collections to further refine the second HMW pentagona DNA extraction to above 15,000bp fragments only
Sample Prep
11/12/19
- Took out electrophoresis buffer, marker, and loading solution from the 4 degree fridge at least 30 minutes before use to get to room temp
- Prepared PCR tubes with all of each sample. This was:
- 27.5µl PG3
- 26µl PG4
- Added 2.5µl and 4µl of TE buffer to each sample to bring up to 30µl each respectively
- Added 10µl of room temp loading solution to each sample and pipetted to mix
- Realized that I couldn’t do both at the same time, so PG4 with loading solution was placed in the 4 degree fridge until PG3 was finished in the Pippin
BluePippin Prep
11/12/19
- Note two half used cassettes were used, meaning two runs each with one sample and one marker were done for these 2 samples
- Brought p20, p200, cassette, tips, solutions, and samples up to the GSC
- Edited the protocol for this cassette:
- Selected “range” for lane 2
- Selected reference for lane 1
- Clicked “apply reference to all lanes”
- Unclicked range and set reference to off for lanes 5, 4, and 3
- Set start as 15,000bp and end as 150,000bp so the target was 82,500bp for lane 1
- Saved protocol as MES_15kb_HighPass
- Calibrated the optics feature
- Unwrapped cassette, removed foil seals I have used to save it from last time, and looked for problems with the gel: none
- Checked amount of buffer in reservoirs: all well full
- Removed all buffer from elution modules (~80µl) and replaced it with fresh electrophoresis buffer 80µl. Do this in all elution modules even ones not using
- Sealed elution modules with sicky provided with the cassettes
- Closed lid and performed continuity test: failed only in lane 5 Previously I had talked to Sage Science technicians about reusing cassettes and they said that if wells/lanes that were used before fail the continuity test it’s fine to use the other lanes that pass and that you haven’t used before
- Removed 40µl of buffer from 2 and 1 sample wells
- Added 40µl marker U1 to well 2
- Added 40µl PG3 to well 1
- Closed lid and pressed start
- Program ran until idle, 2.5 hours later
- Set timer for 45 min to let all sample elute and waited to remove until after that
- After 45 min, removed sticker and saved all the liquid in elution modules 1
- Filled elution modules 1 with 80µl 0.1% Tween and waited 1 minute, then removed and saved that liquid as well
- Put those samples in the 4 degree fridge
- Followed the exact same steps above with the second half used cassette and with sample PG4
QC
- High sensitivity Qubit of top and bottom of samples and tween washes, after gently flicking to mix
| Sample | Standard 1 | Standard 2 | Reading 1 ng/µl | Reading 2 ng/µl | Average ng/µl |
|---|---|---|---|---|---|
| PG3 Top | 47 | 24579 | 16.5 | 16.5 | 16.5 |
| PG3 Bottom | 47 | 24579 | 19.6 | 19.6 | 19.6 |
| PG4 Top | 47 | 24579 | 34.2 | 34.2 | 34.2 |
| PG4 Bottom | 47 | 24579 | 33.6 | 33.8 | 33.7 |
| PG3 Tween Top | 47 | 24579 | 1.56 | 1.58 | 1.57 |
| PG3 Tween Bottom | 47 | 24579 | 1.71 | 1.73 | 1.72 |
| PG4 Tween Top | 47 | 24579 | 0.47 | 0.478 | 0.474 |
| PG4 Tween Bottom | 47 | 24579 | 0.516 | 0.524 | 0.54 |
- Ran D5000 tapestation on the elution module liquid (not the tween washes)
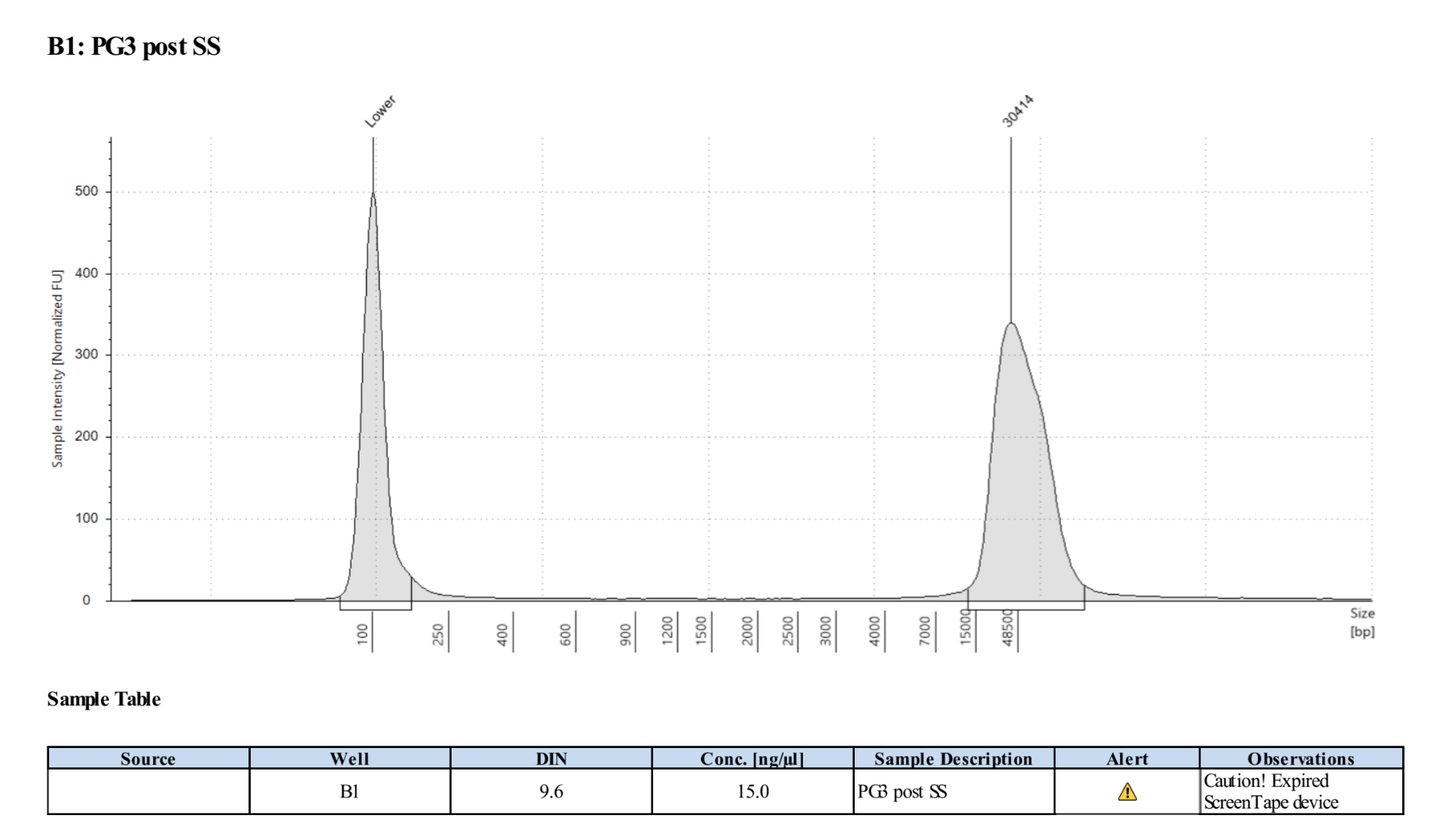
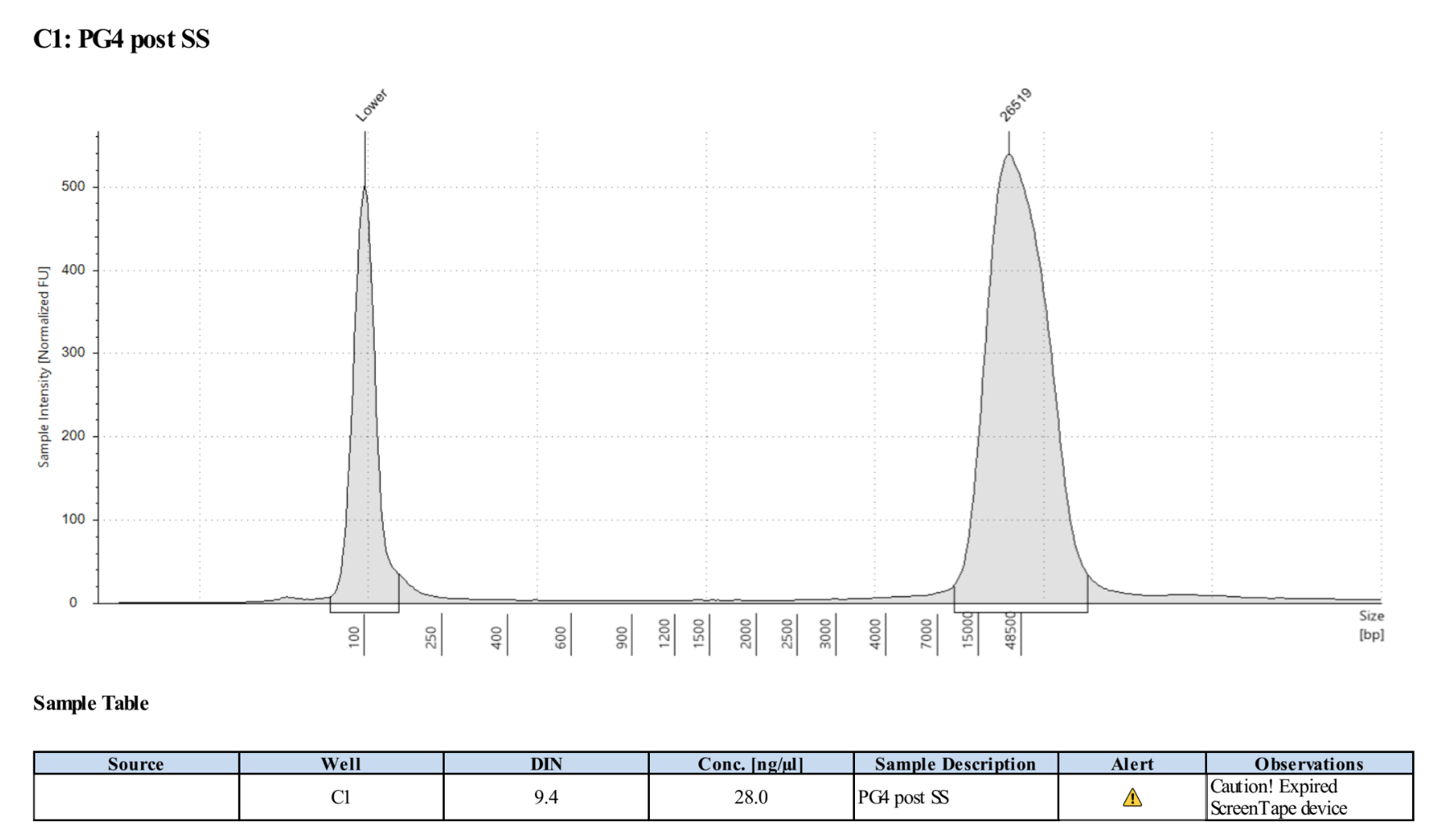
0.45X Cleanup of Elution Liquid and Tween Washes
11/13/19 and 11/14/19
- Made fresh 80% EtOH before starting
- Took KAPA Pure Beads out of fridge about 30 minutes before they were needed
- Used pipettes to determine how much volume of sample out of the BluePippin there was:
- PG3: 73µl so .45X is 32.85µl of KAPA Pure Beads
- PG4: 74µl so .45X is 33.3µl of KAPA Pure Beads
- PG3T: 78µl so .45X is 35.1µl of KAPA Pure Beads
- PG4T: 80µl so .45X is 36µl of KAPA Pure Beads
- Added the above amount of beads to each sample very gently then flicked the tubes gently until a homogeneous distribution of beads was obtained and then tubes were spun down briefly in a minifuge
- Placed tubes on shaker for 20 minutes at room temp at 300rpm
- Then placed the tubes on the magnet rack and waited 10 minutes
- Removed the clear supernatant and SAVED as the sample name and “supernatant”
- Added 200µl of fresh 80% EtOH to each tube gently then waited 30 seconds
- Removed and SAVED as the sample name and “wash 1”
- Added 200µl of fresh 80% EtOH to each tube gently then waited 30 seconds
- Removed and SAVED as the sample name and “wash 2”
- Spun sample tubes briefly in tabletop minifuge and placed back on the magnet rack
- Used p20 to get the last liquid out of the tube, just a few µl
- Did not let samples rest really, removed tubes from rack and added 50µl of nuclease free water to each tube by gently dripping
- Flicked tubes until the pellets resuspended and then a little further, looked at beads very closely and tried to flick away any small clumps, and then briefly spun down because liquid was all on the lids
- Placed tubes closed on sample rack still at room temp for 20 minutes
- After then placed on shaker at room temp for afternoon and overnight at 300prm
- The next morning, spun down tubes briefly because some evaporation, also the bead distribution was different between the real samples and the washes:
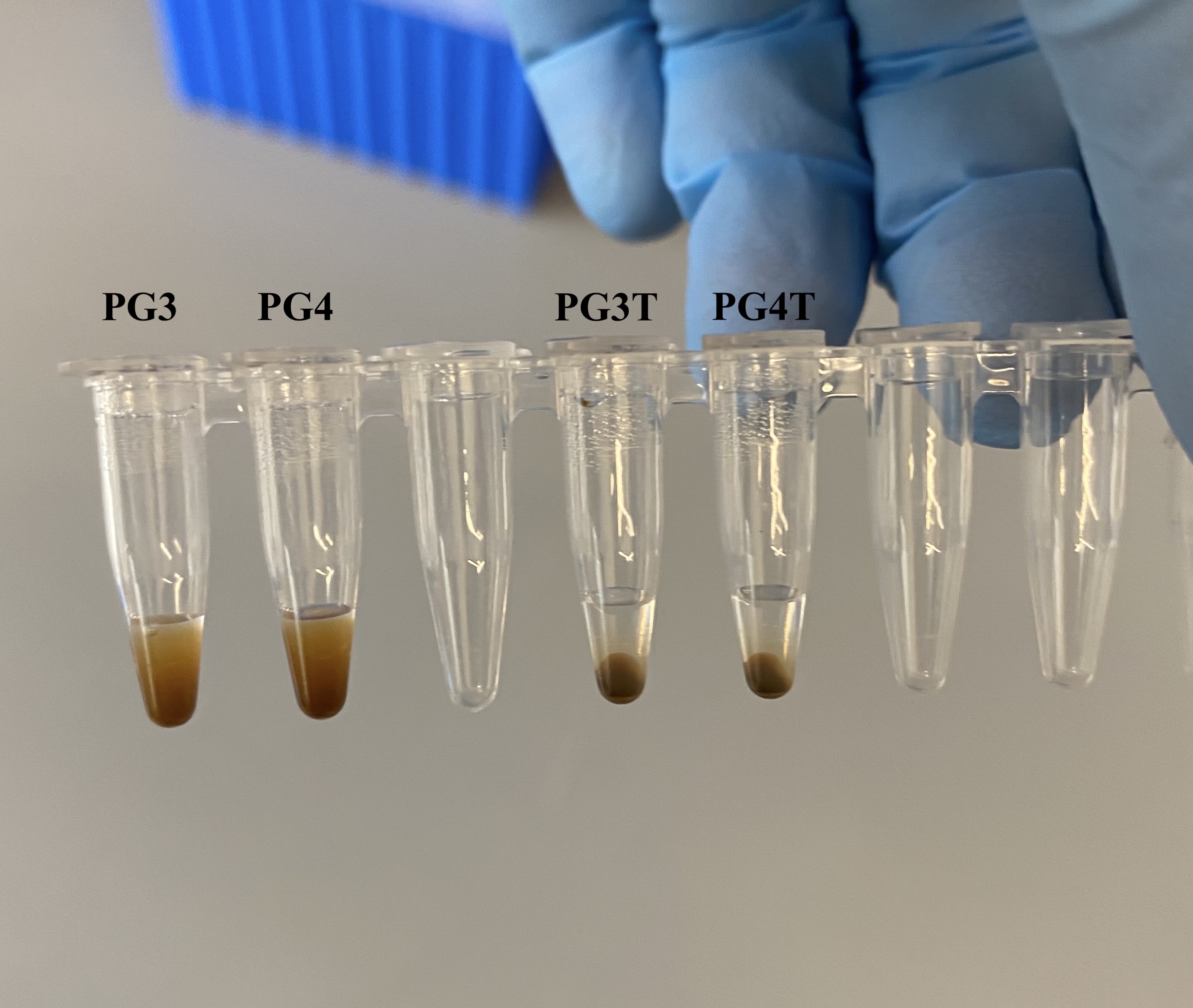
- Placed tubes on the magnet rack for 10 minutes
- Removed the clear supernatant and placed into new labeled sample tubes
- Added 50µl of nuclease-free water to the beads again and saved in 4 degree fridge
High Sensitivity Qubit of top and bottom of sample tubes and washes
Flipped tube end over end to try to mix sample before taking 1µl each to Qubit
| Sample | Standard 1 | Standard 2 | Reading 1 ng/µl | Reading 2 ng/µl | Average ng/µl |
|---|---|---|---|---|---|
| PG3 Top | 41.58 | 24660 | 16.3 | 16.3 | 16.3 |
| PG3 Bottom | 41.58 | 24660 | 17.1 | 17.1 | 17.1 |
| PG4 Top | 41.58 | 24660 | 33.4 | 33.6 | 33.5 |
| PG4 Bottom | 41.58 | 24660 | 30.6 | 30.6 | 30.6 |
| PG3 Tween Top | 41.58 | 24660 | 0.69 | 0.656 | 0.67 |
| PG3 Tween Bottom | 41.58 | 24660 | 0.656 | 0.662 | 0.658 |
| PG4 Tween Top | 41.58 | 24660 | 0.182 | 0.19 | 0.186 |
| PG4 Tween Bottom | 41.58 | 24660 | 0.178 | 0.182 | 0.18 |
Written on November 12, 2019