More HMW Extractions on pentagona seastar with Zymo Quick-DNA HMW MagBead Kit
Using the Zymo Quick-DNA HMW MagBead Kit with the rest of one Cryptasterina pentagona tissue sample in DNA/RNA shield to try to get maximum HMW DNA!!
Sample Prep
- Thawed PG sample from the previous extraction
- Prepped two 1.5mL DNA lo-bind tubes, each with 95µl DNA elution buffer, 95µl Biofluid and Solid Tissue buffer, and 10µl Proteinase K
- Took out the piece of tissue, cut it in half on foil, then chopped with the razor blade until tenderized. Put one 1/2 piece of tissue in each prepared tube.
Whole tissue chunk: Tenderized:
Tenderized:

- Pipetted to mix with p200 pipette
- Placed in thermomixer, 55 degrees C and shaking at 500rpm. Put in at 4:30pm and planned to go overnight
- Samples were looked at the next morning at 10am, some tissue chunk was still visible, so they were vortexed for 10 seconds each, then returned to the thermomixer for another 45 minutes
- Samples were taken out of the thermomixer and centrifuged at maximum speed (20,000rcf +) for 1 minute
- About 200µl of supernatant was pipetted off into new lo-bind tubes, mostly no chunks were transferred
DNA Purification
- Added 400µl of Quick-DNA MagBinding Buffer to each sample and pipetted to mix
- Gently shook/swirled MagBinding Beads to make sure they were resuspended
- Added 33µl MagBinding Beads to each sample, making sure to shake the bottle of beads before aspirating from it each time
- Pipetted to mis the beads inside the sample tubes
- Samples were incubated for 15 minutes at 12,000rpm on the thermomixer at 23 degrees
- Placed the tubes on the magnet rack an waited until the beads went to the magnet. note: the liquid never went clear, but it was yellowish so I thought it was the tissue remnant not the beads because the beads are opaque black
- Carefully removed and discarded supernatant
- Took tubes of the magnet and added 100µl DNA Elution buffer to each sample and pipetted to mix
- Added 500µl of Quick-DNA MagBinding Buffer to each tube and pipetted up and down to mix
- Placed on the thermomixer 1200rpm for 10 min at 23 degrees C
- Took out samples and placed on the magnet rack
- When the supernatant was clear, removed and discarded it from each tube
- Took samples off the magnet and resuspended the beads in 500µl DNA Pre-wash Buffer and pipetted 10 times to mix
- Placed the tubes back on the magnet stand and removed the supernatant when it was clear
- Took the tubes off the magnet and resuspended the beads in 900µl g-DNA Wash Buffer and pipetted 10 times to mix
- Removed entire volume of each sample including the beads and placed into new 1.5mL lo-bind tubes
- Placed tubes on magnet and removed the supernatant when it was clear
- Took tubes off the magnet and resuspended the beads in 900µl g-DNA Wash Buffer and pipetted 10 times to mix
- Removed the entire volume of each sample including the beads and put into new 1.5mL lo-bind tubes
- Placed tubes on magnet stand and removed the supernatant when it was clear, used a smaller pipette to get rid of any excess liquid I could see in the tube
- Left the tubes on the magnet stand with their tops open for ~20 minutes. The top of the beads were nice and matte but the bottom was still a little bit damp looking, but no beads were looking glossy anymore. For PG3 it was sooner than PG4 by about 5 minutes
- Resuspended the beads in 33µl TE buffer from the kit and pipetted at least 20 times to mix
- Placed the tubes on the thermomixer at 23 degrees C 1200 rpm shaking at 12:30am.
- Tubes were taken off the shaker at 7pm and placed on the magnet rack
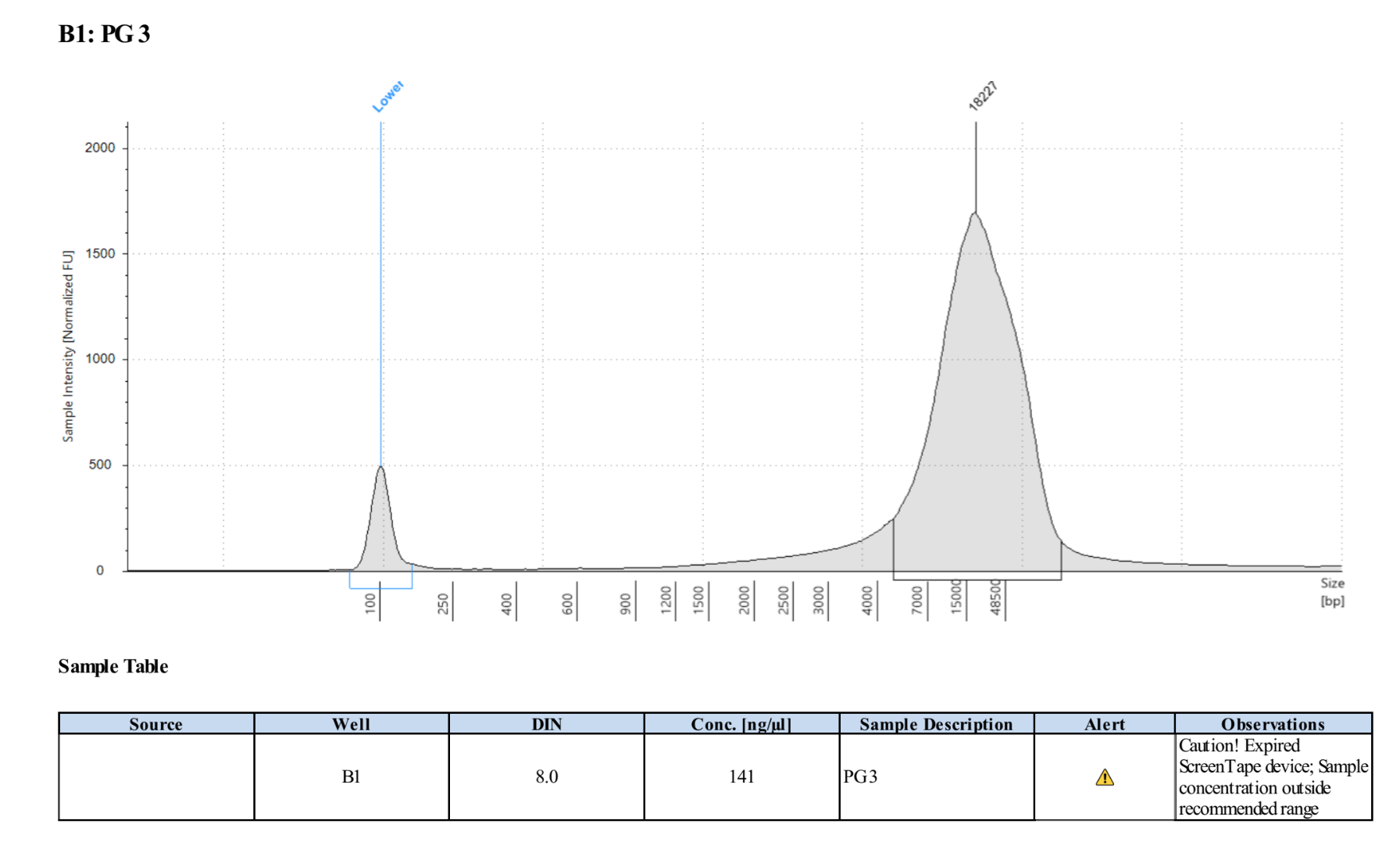
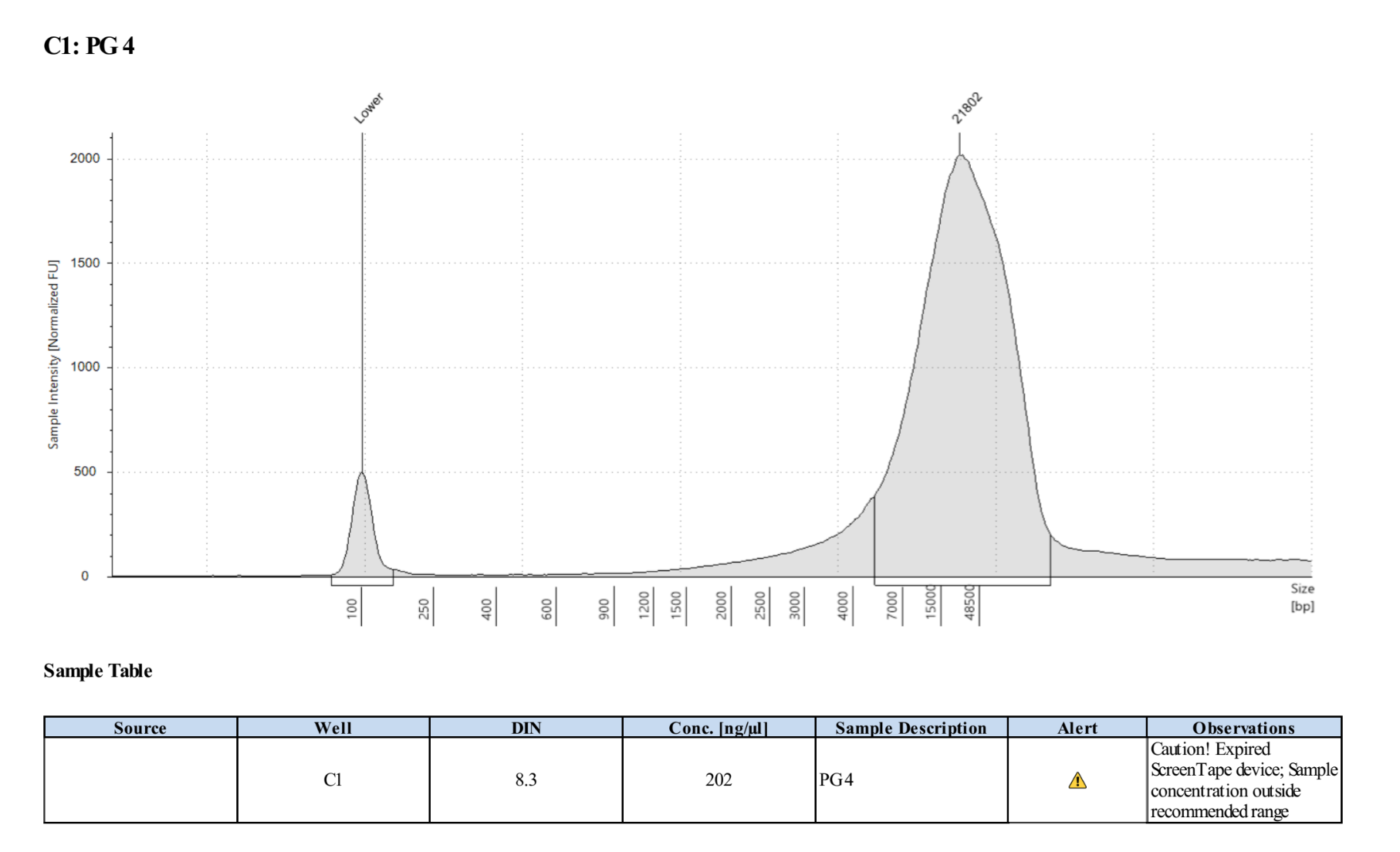
- The clear supernatant was taken and SAVED as the DNA! 2µl was used for Qubit and 1µl for Tapestation that same day, then the samples were placed in the 4 degree fridge
Broad Range Qubit:
Samples were flicked gently before and a top and bottom 1µl were quantified
Standard 1: 190 RFU
Standard 2: 230434 RFU
PG3 Top: 164ng/µl, 161ng/µl, average: 162.5ng/µl
PG3 Bottom: 161ng/µl, 159ng/µl, average: 160ng/µl
PG4 Top: 220ng/µl, 218ng/µl, average: 219ng/µl
PG4 Bottom: 218ng/µl, 216ng/µl, average: 217ng/µl
Written on November 10, 2019