Testing Zymo Quick-DNA HMW MagBead Kit on pentagona Seastar Tissue
Testing the Zymo Quick-DNA HMW MagBead Kit with one Cryptasterina pentagona tissue sample in DNA/RNA shield
Sample Prep
- Added 260µl of Pro k storage buffer to the 5mg of powder Pro k provided with the kit, then kept on ice
- Thawed one PG sample on ice
- Prepped two 1.5mL tubes, each with 95µl DNA elution buffer, 95µl Biofluid and Solid Tissue buffer, and 10µl Proteinase K
- Took out the piece of tissue, cut it in half on foil, then cut one half in half again. Put one 1/4th piece of tissue in each prepared tube. Placed the rest of the tissue back in the tube with the DNA/RNA shield and re-froze at -20
- Pipetted to mix with p200 pipette
- Placed in thermomixer, 55 degrees C and shaking at 400rpm. Put in at 3:39pm and planned to go overnight
- Samples were taken out of the thermomixer at 9:15am the next day. note: samples weren’t completely broken down, there was some tissue bits left around the bottom of the tubes. PG1 had what I would guess were fat bits floating at the top but those weren’t in PG2
- Centrifuged tubes at 10,000 rcf for 1 minute
- Tried to transfer only liquid to new 1.5mL tubes, basically failed at that even with the p20 pipette, there was still some tissue, or more likely mucus/lipids that came to the next tube
- Centrifuged again now at max rcf for 2 minutes
- Transferred what liquid I could without getting any chunks to new 1.5mL tubes again. PG1 had a few floating mucus/lipid bits that I could not avoid pipetting. PG2 had a top orange layer in the liquid that I also transferred
DNA Purification
- Added 400µl of Quick-DNA MagBinding Buffer to each sample and pipetted to mix this addition seemed to remove/dissolve any mucus-y-ness that was left in the samples
¯\_(ツ)_/¯ - Gently shook/swirled MagBinding Beads to make sure they were resuspended
- Added 33µl MagBinding Beads to each sample, making sure to shake the bottle of beads before aspirating from it each time
- Pipetted to mis the beads inside the sample tubes
- Placed on the shaker for 15 minutes, the protocol said 1100 rpm, but I thought that was really fast and I thought the tubes were going to come of the shaker, so I did 740 rpm. However, I came back after 15 minutes and saw that some of the beads had settled so I switched the tubes to the thermomixer at 23 degrees C and 12,000 rpm for 10 minutes. This time the beads were resuspended
- Placed the tubes on the magnet rack an waited until the beads went to the magnet. note: the liquid never went clear, but it was yellowish so I thought it was the tissue remnant not the beads because the beads are opaque black
- Carefully removed and discarded supernatant
- Took tubes of the magnet and added 100µl DNA Elution buffer to each sample and pipetted to mix note: PG1 had what was like a clump of beads that wouldn’t really breakup, but I was being gentle and din’t try very hard also
- Added 500µl of Quick-DNA MagBinding Buffer to each tube and pipetted up and down to mix (still saw the clump in PG1)
- Placed on the thermomixer 1200rpm for 10 min at 23 degrees C
- Took out samples and placed on the magnet rack
- When the supernatant was clear, removed and discarded it from each tube
- Took samples off the magnet and resuspended the beads in 500µl DNA Pre-wash Buffer and pipetted 10 times to mix
- Placed the tubes back on the magnet stand and removed the supernatant when it was clear
- Took the tubes off the magnet and resuspended the beads in 900µl g-DNA Wash Buffer and pipetted 10 times to mix
- Removed entire volume of each sample including the beads and placed into new 1.5mL tubes
- Placed tubes on magnet and removed the supernatant when it was clear
- Took tubes off the magnet and resuspended the beads in 900µl g-DNA Wash Buffer and pipetted 10 times to mix
- Removed the entire volume of each sample including the beads and put into new 1.5mL tubes
- Placed tubes on magnet stand and removed the supernatant when it was clear, used a smaller pipette to get rid of any excess liquid I could see in the tube
- Left the tubes on the magnet stand with their tops open for ~25 minutes. The top of the beads were nice and matte but the bottom was still a little bit damp looking, but no beads were looking glossy anymore. Did not want to over dry, at first I set a timer for only 15 minutes but the beads were very wet then still
- Resuspended the beads in 50µl DNA elution buffer from the kit and pipetted at least 20 times to mix
- Placed the tubes on the thermomixer at 23 degrees C 1200 rpm shaking at 11:32am.
- Tubes were taken off the shaker at 4:15pm and placed on the magnet rack
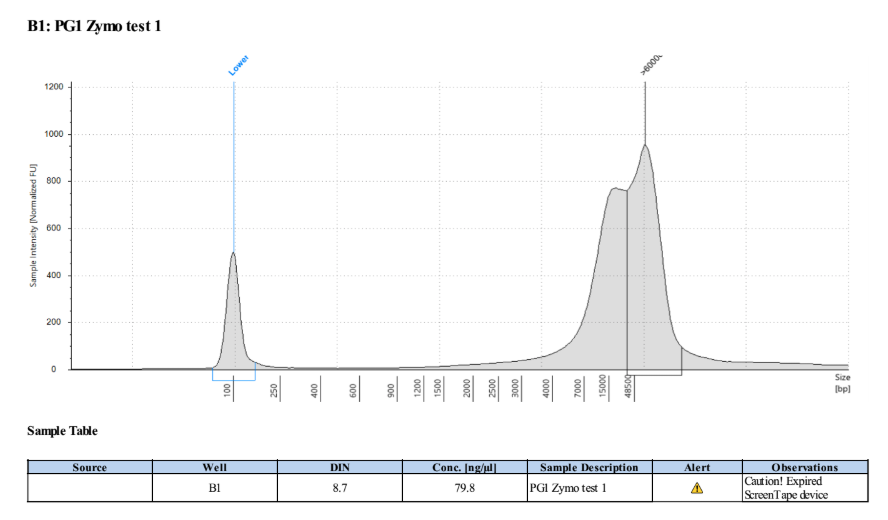
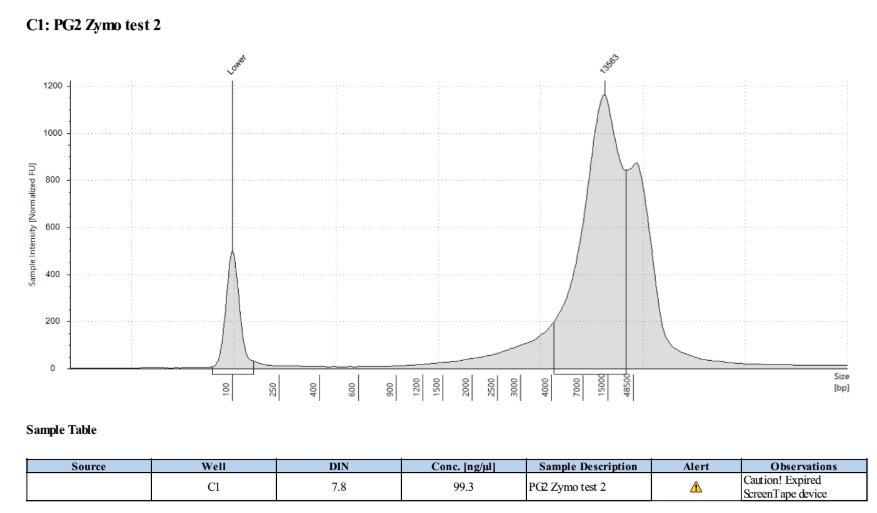
- The clear supernatant was taken and SAVED as the DNA! 1µl was used for Qubit and Tapestation each, the rest was put into the -20 freezer
Broad Range Qubit:
Standard 1: 200 RFU
Standard 2: 23234 RFU
PG1: 96.2ng/µl, 95.8ng/µl, average: 96ng/µl
PG2: 127ng/µl, 126ng/µl, averageL 126.5ng/µl
Written on October 31, 2019