PocHistone Amplification and Sanger Sequencing Prep Protocol
Protocol for PocHistone Amplification and Sanger Sequencing for Determining Pocillopora Species
Goal:
Amplify the histone 3 region region of Pocillopora coral DNA using PCR (polymerase chain reaction). Then use amplification product for Sanger Sequencing and determination of Pocillopora coral species between P. eydouxi and P. meandrina. Primer sequences and concept developed in Johnston et. al 2018.
Information on Sanger sequencing at the URI GSC is here. Sequencing takes place after 10am on Tuesdays and Thursdays.
Materials and Equipment:
- Primers (oderable from IDT):
- PocHistoneF: 5′-ATTCAGTCTCACTCACTCACTCAC-3’
- PocHistoneR: 5′-TATCTTCGAACAGACCCACCAAAT-3′
- Phusion High-Fidelity PCR Master Mix with HF Buffer
- Ultrapure free water/PCR grade water
- Thermocylers (Eppendorf Mastercycler Pro, up to 3)
- 96 well plates and foil seals
- Microcentrifuge tubes
- 5mL tubes
- Multichannel and regular micropipettes
- Filter pipette tips
- Ethanol
- Magnetic DNA binding beads
- Magnetic plate for 96 well plates
- Materials for gel electrophoresis (agarose, TAE buffer, gel box, gelgreen, 1KB plus DNA ladder, loading dye)
- Broad range dsDNA Qubit assay and tubes for Qubit use
Protocol Steps
- Primer working stock dilution
- DNA dilution
- PocHistone amplification
- 1X bead clean up
- Qubit and Gel
- DNA dilution and prep for sequencing
Primer Working Stock Dilution
- Primers are resuspended to 200uM stock solution (they come dried) with low TE buffer
- Primers should be added at 10uM concentration for each, and thus need to be diluted into workin stock aliquots
- For working stocks of 500ul in 1.5mL tubes:
- 25ul of 200uM Stock Primer
- 475ul of UltraPure water
- Vortex to mix for 15 seconds then spin down
- Store in -20 degree C freezer
DNA Dilution
- Dilute all samples to 10ng/ul in 10, 20, or 25ul
- For a lot of samples (20+) I recommend making dilutions in a plate, so they can be easily added to reaction plates in the exact same order with a mulichannel pipette. You can even add in wells with just water as negative controls to the dilution plate, so that the “water sample” gets multichannel pipetted at the same time as your sample DNA. I also recommend making plate maps for dilutions, one for the order of your samples, one for the amount of DNA to add, and one for the amount of water to add. DNA 1.5mL tubes can be organized in the exact order of the plate on a large black rack (96 spots) as well for visual ease. See example below of plate layout and sample layout for a dilution plate of 32 samples and 3 negative controls (just water). This was printed out and each cell was highlighted when the amount of liquid was added.
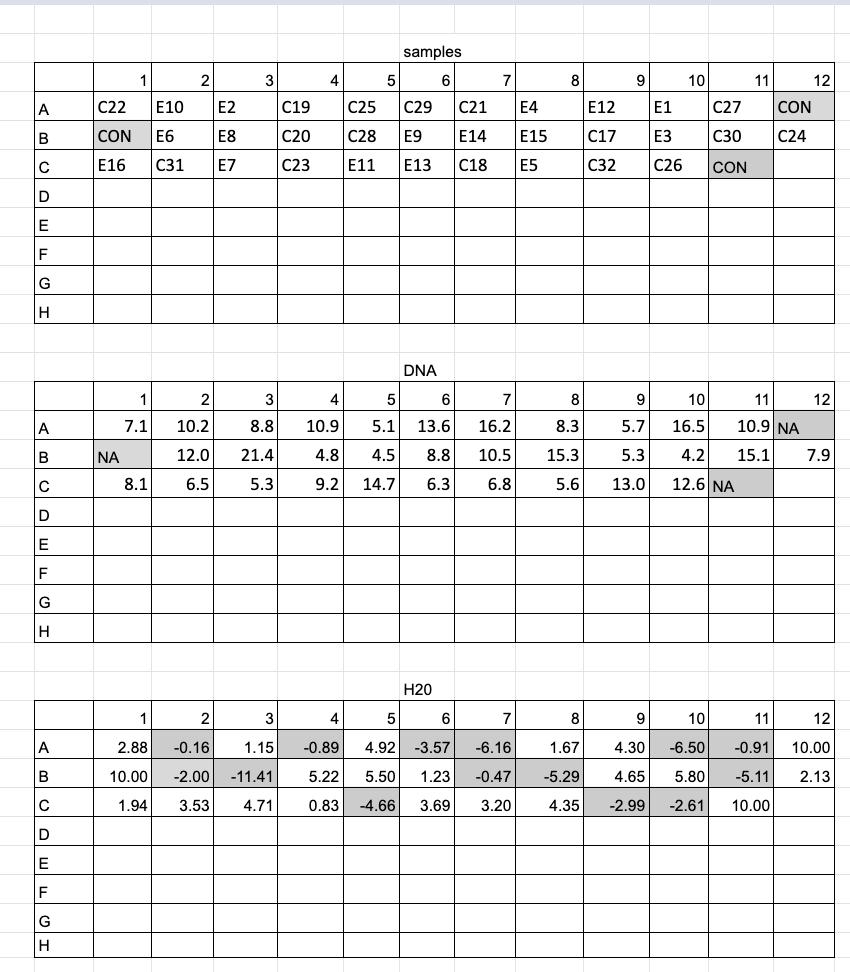
- However many microliters you chose to do the dilution in depends some on the concentration of DNA you start with. As you can see above many of the samples had less than 10ng/ul as their starting concentration. Those samples were just used as is. And the low concentrations was why I chose to dilute in 10ul total to not use too much DNA.
- Add the planned ng of DNA to their planned well in a plate and the planned ul of UltraPure water to their wells
- Do either all DNA or all water at a time. It’s always best practice to add the larger volume liquid to an empty well first
- Seal plate with sticky foil, vortex, and spin down in the large centrifuge
- Keep plate on ice if using the same day, freezer if not
PocHistone Amplification
- Each sample will be amplified one time in a 33ul reaction volume
- Calculate the amount of master mix is needed: add number of samples and negative controls plus a % error (5-10%) to get the n number
- Make master mix in 1.5mL or 5mL tube with these components scaled up to your n number:
- 16.66ul Phusion PCR master mix * n =
- 14.66ul UltraPure water * n =
- 0.43ul 10uM working stock FatP6.1 primer * n =
- 0.43ul 10uM working stock RORF primer * n =
- Vortex and spin down master mix and keep on ice
- Use a new 96 well plate and add 32ul of master mix to as many wells as samples you have + the number of negative controls planned (usually 1 per row). For ease of pipetting, this is the same layout of the dilution plate you already made
- Using a p2 multichannel pipette, add 1ul of DNA sample to their respective well in the plate where the master mix is added. Be careful to keep orientation of plates the same and of the multichannel the same when transferring between the two plates. If you don’t already have aliquots for negative controls, add 1ul of water in the wells for those reactions
- Cover plate with foil seal and vortex to mix. Centrifuge down to remove bubbles
- You will need 1 thermocycler (if you have more than 96 samples you’ll need more than 1)
- Turn on thermocyler and login to PUTNAM (1,2,3,4) and navigate to the PocHistone program
- Run that program for however many plates you have
- Program, bold fields are cycled 30 times:
- 60 seconds 94 degrees C
- 30 seconds 94 degrees C
- 30 seconds 53 degrees C
- 60 seconds 72 degrees C
- 5 minutes 72 degrees C
- The program runs for about 1 hour and 40 minutes
- Take plates out and store at 4 degrees C if not cleaning up immediately
1X Bead Cleanup (post PCR bench)
- Take KAPA pure beads out of 4 degree fridge ~30 minutes before use to get to room temperature, and swirl to even out the beads (which settle on the bottom), and keep them in a drawer while warming out of the light
- Make fresh 80% ethanol in a 50mL conical for the day, pour out 40mL of 100% ethanol into the conical, and add ultrapure water up to 50mL
- Take plates out of either fridge or thermocyler and spin down in centrifuge
- You can either add beads 1 well at a time or by row using a multichannel and a liquid trough/basin. If you have more than 16 samples, it saves time to use a trough
- Take beads out of the drawer and swirl again. If using a trough, calculate how many beads you’ll need: 33ul * n and pipette that volume of beads into the trough
- Adding beads to samples: If adding to individual wells, us a p200 and add 33ul of beads to each well, slowly because the bead liquid is very viscus. Pipette up and down at least 10 times per sample to mix, pipette slowly to avoid bubbles forming. You have to completely mix gently here, your DNA is binding to the beads in this step. The same process applies to using the multichannel, suck up 33ul with all the tips and slowly add to your row in the plate, pipette mixing at least 10 times slowly. Repeat for all rows/samples, avoid bubbles because they can effect magnetic binding, tap down plate gently if some appear
- Take the rotating shaker off the shared equipment shelf and plug in by the qubit. Let your bead-sample plate incubate shaking for 15 minutes at 200 speed to facilitate DNA binding
- After incubation, place the plate on the magnet plate (white plate with silver metal knobs), and place back on the shaker for ~5 minutes. Larger volumes can take longer for the beads to magnetize so shaking helps
- Take plate back to bench. Set up a trough for “supernatant”, a trough for EtOH, and for ultra pure water
- Pour some EtOH and water into their troughs but leave the supernatant one empty
- When the liquid in the wells has gone clear, carefully use the multichannel to pipette out the clear supernatant without disturbing the beads (set pipettes to ~55ul), it helps to slide the tips down the sides of the tubes without the beads. The clear sides alternates each row. Discard supernatant in the labeled trough and change tips. Remove supernatant from every well
- Add 100ul of 80% EtOH to each well carefully without disturbing the beads, again you can put the tips on the side of the wells without the beads
- Remove the clear supernatant from each well without disturbing the beads and discard into the supernatant trough, changing tips for each well
- Repeat the last two steps (2 ethanol washes)
- Use the p20 multichannel pipette to suck up any remaining ethanol at the bottom of the wells of the plate (still on magnet)
- Wait ~2 minutes to let any residual ethanol to dry
- Take plate off the magnet into a regular rack
- Set multichannel to 30ul and add 30ul of ultrapure nuclease-free water to each well of the plate. Carefully add the water to the beads, you’ll have to pipette up and down slowly as long as it takes to resuspend the beads off the side of the well. Alternate the tip position to get beads stuck on opposite sides of the wells. Avoid making bubbles and repeat for every row
- Place plate on the shaker at 200 rpm for 5 minutes
- Get another plate ready for the cleaned amplifications
- Once done shaking, place the plate back on the magnet and wait until the wells become clear
- Remove the 30ul of clear supernatant from each well and into the new 96-well plate
- Seal the plate with a labeled foil seal and place in the fridge if quantifying the next day, or freezer if it will be there over the weekend
Qubit or Plate Reader Assay and gel
- Use the broad range dsDNA Qubit assay to quantify every one of the amplified and cleaned samples, or if doing more than ~30, use the plate reader
- Qubit protocol
- If you have over 20-30 samples, you can use the plate reader to quantify your DNA using the Biotium PicoGreen Assay
- Plate Reader Protocol
- Run a 1% gel on all of the amplified and cleaned samples, use a 1kb plus DNA ladder. There should be 1 band at ~670bp. Other bands are signs of potential contamination or off target amplification
- Gel protocol
Prep for sequencing (post PCR bench)
Notes
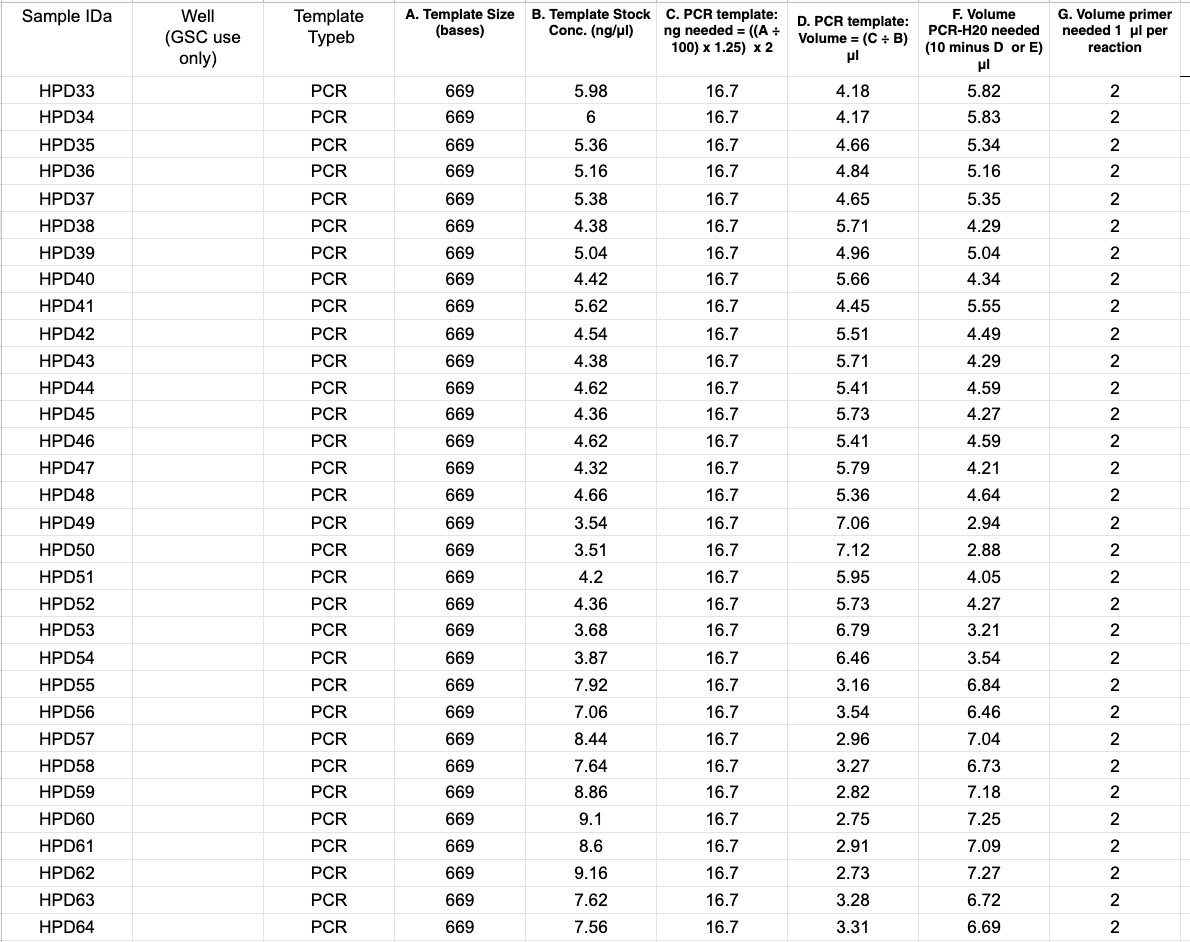
- For Sanger sequencing at the URI GSC the amount of DNA to send them depends on how long the fragment is. The equation for how many ng to send is
((number of bases / 100)) * 1.25) * 2
For PocHistone the calculation is: ((669/100)* 1.25) * 2 = 16.7. That means you’ll need 16.7ng of each amplification - 16.7 ng is not a lot of DNA, so some of your Qubit values are likely going to be higher than 16.7ng/ul. It’s best practice not to pipette below 1ul, so it’s probably you’ll need to dilute the amplifications
- The volume of liquid sent to the GSC is 12ul total, and 2ul of that is one of the primers. So you have 10ul to use for DNA. Depending on the concentrations, you’ll have to decide if a 1:10, 1:5, 1:2, etc dilution will work to pipette over 1ul for 16.7ng but under 10ul
- You’ll also have to dilute one of the primers to send with your DNA. Sanger sequencing amplifies the fragment with one of the two primers. Make sure to use the reverse primer for PocHistone. Primers are at a working stock concentration of 10uM, but need to be added as 2ul of 3.2uM concentration to the 10ul of DNA
Steps for sequencing prep:
- Dilute DNA by however much you need. At this time you’ll probably make a new plate, so don’t include the negative control reactions. As an example, to do this you could use a multichannel to take 2ul from each sample into a new plate, and add 8ul of nuclease free water to each well. That would be a 1:5 dilution
- Dilute the PocHistone R primer to 3.2uM in volume for 2ul per sample. Calculate # of samples * 2 to get the volume. You can use this solution dilution calculator to calculate how to dilute the primer in that volume
- Calculate volume of DNA for 16.7ng for each sample (use the dilution factor you used on your previous qubit values)
- Aliquot plate of DNA (not including negative controls) with 16.7ng of DNA per sample and nuclease free water up to 10ul for each well. If you did a dilution on your sample, use this. Using a plate map here where you highlight off each well after adding the right amount is very helpful. Each well will have a different amount of DNA and water. Also make sure the samples go in the order you have planned because plates do not have room for labels

- Add 2ul of 3.2uM reverse primer to each well and cover with a foil seal. Label the seal with something like “Putnam Lab samples for Sanger sequencing, X number of samples, date, initials etc”
- Prepare a spreadsheet for GSC with all of this information. You can use the document provided by the GSC link, or you can make your own spreadsheet and attach it in an email to the GSC (see below). It’s basically all the calculations you did for the correct amount of DNA for the template, so the GSC knows how much DNA there is and the fragment length. Email this sheet to the GSC at jatoyan@uri.edu or gsc@etal.uri.edu saying you are submitting how ever many samples for Sanger sequencing for the Putnam Lab using her blanket PO (purchase order). Include the label on top of the plate that you wrote. Notice the Sample ID column for the GSC uses a sample code that is 3 letters then a number, which makes it easier for the GSC. Make sure you know what GSC sample ID matches with your sample numbers
- Take plate/tubes up to the GSC and place in freezer next to the door. There is a place on the top shelf that with a label that says “samples for GSC sequencing”, place your tubes or plate in there or near that rack
You should get the sequences back the next day!
Written on January 25, 2021