Second Test PCR for mtORF Pocillopra Species Determination
Testing Amplification of Danielle’s Pocillopora Samples and Reduced Cycle Number for mtORF Amplification
Goal: See if reducing cycle number gives a clearer amplification band. Also want to see if Danielle’s samples will amplify
Major Results: The PCRs look the same, and Danielle’s samples amplified well
Major Takeaways: Cycle number doesn’t change the size smearing, I sent the gel to Janet and she says it could be too much DNA in the gel
Samples Used: E2, E6, E10, 65, and 190
Decreased cycles in program to 30, and planned to follow more closely the triplicate 33ul reactions in the ITS2 protocol instead of the 100ul reactions
- Made a master mix for 100ul for each sample, which will then be split up into 3 reactions
- 50ul phusion master mix * 6.3 = 315ul
- 1.3ul FatP6.1 10uM * 6.3 = 8.19ul
- 1.3ul RORF 10uM * 6.3 = 8.19ul
- 44.4ul ultra pure water * 6.3 = 279.72ul
- Aliquoted 97ul of master mix into 6 wells of a PCR strip tube labeled for each sample and a control
- Added 3ul of DNA from each sample to their appropriate tube
- Added 3ul of nuclease free water to the control tube
- Vortexed and spun down tubes
- Made 3 other sets of strip tubes for triplicate reactions and labeled with sample numbers and control
- Used the multichannel to add 33ul of mix+DNA to each samples’ well
- Spun down strip tubes and placed in FATP6 RORF PCR program
- Program ran for 1 hour ~39 minutes
Gel
- Followed protocol for making a 1% small gel
- Used 1kb plus generuler as ladder
- Ran gel for 60 minutes at 100volts
- The last three samples are from a Puritz lab project that I ran because there was space on this gel
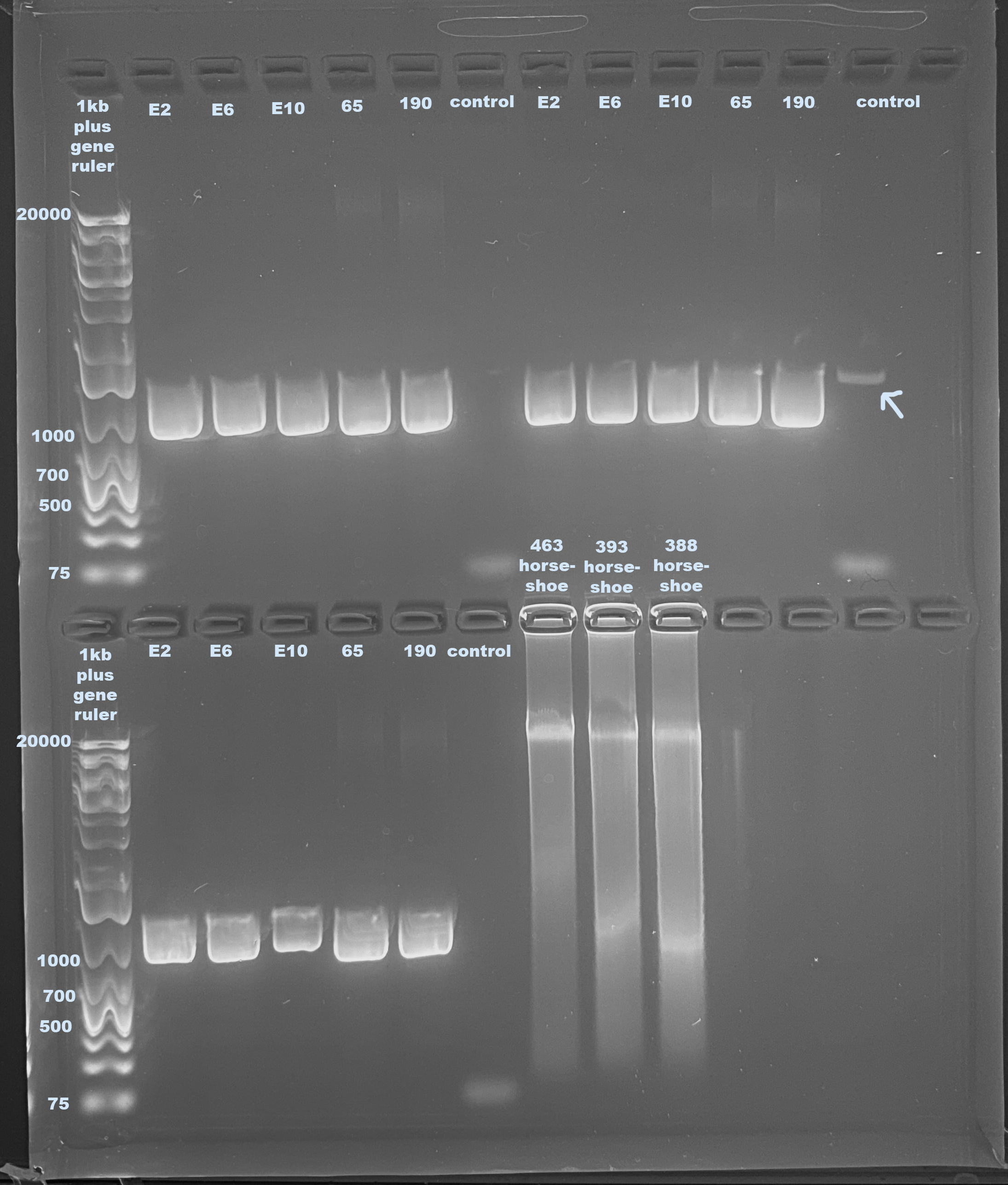
Danielle’s samples look good, they amplified in the same way. However the same smear band is present. I emailed Janet about this and she said it could be that there was too much DNA in the get input. I have not yet qubited these samples so that could be true. There also looks like there is a tiny amplification in one of the controls, which is interesting. I don’t know how it could be in one and not the others because it was from the same stock.
Next steps are to bead clean, quantify, and aliquot for test sequencing. Janet said that would be a clear way to see if I am amplifying two things.