Oyster Nutrient Enrichment Treatment RNA Lib Prep with Zymo-Seq RiboFree Total RNA Library Prep Kit
We have more free RNA Library Prep Kit Sequencing, using nutrient enriched and control treated eastern oysters (Crassostrea virginica) from Rebecca Stevick using the Zymo-Seq RiboFree Total RNA Library Prep Kit
RNA samples were given to me each with 300ng and in 8µl volume. These are the numbers on each tube, and what nutrient enrichment treatment they received, and what experimental bucket they were in. Tube number was used moving forward to reduce confusion.
| Tube Number | Treatment | Sample Name | Bucket |
|---|---|---|---|
| 13 | Control | BC.7.1 | 7 |
| 14 | Control | BC.7.2 | 7 |
| 15 | Control | BC.7.3 | 7 |
| 16 | Control | BC.9.1 | 9 |
| 17 | Control | BC.9.2 | 9 |
| 18 | Control | BC.9.3 | 9 |
| 19 | Enriched | BC.11.1 | 11 |
| 20 | Enriched | BC.11.2 | 11 |
| 21 | Enriched | BC.11.3 | 11 |
| 22 | Enriched | BC.12.1 | 12 |
| 23 | Enriched | BC.12.2 | 12 |
| 24 | Enriched | BC.12.3 | 12 |
First Strand cDNA Synthesis
- Thaw samples and reagents R1 and R2 on ice (reagents are kept in the -80)
- Added 2µl reagent R1 to each tube, pipetted to mix and spun down
- Placed in thermocycler MES login and used the “ZRibo 1st Strand” program
- 98°C 3 minutes
- 4°C hold
- 25°C 5 minutes
- 48°C 15 minutes
- 4°C hold
- At the first 4°C hold, added 10µl R2 reagent tp each sample in the thermocycler and pipetted to mix
- Pressed enter in the program to continue it
- Took out reagents D1, D2, and D3 to thaw on ice
RiboFree Universal Depletion
- Added 10µl of reagent D1 to each sample on ice, pipetted to mix and spun down
- Placed tubes in thermocycler MES login ZRibo Depletion program:
- 98°C 3 minutes
- 68°C 5 minutes
- 68°C hold
- 68°C 1 hours this is variable in the protocol, I chose 1 hours because it is for over 250ng but below 1µg input
- 68°C hold
- 98°C 2 minutes
- 25°C hold
- At first hold in the thermocyler program added 10µl D2 reagent in the thermocyler without removing the tubes and pipetted to mix
- Pressed enter in the program to continue it
- At the second hold in the thermocycler program added 10µl of the D3 stop solution to each tube in the thermocyler without removing the tubes and pipetted to mix
- Pressed enter in the program to continue it
- Made a 1.5mL tube of 95% EtOH with ultra pure water
- Once the program was done added 25µl of 95% EtOH to each sample, pipetted to mix, and spun down
Cleanup with DNA MagBeads
- Added 24mL of 100% EtOH to the concentrated wash buffer before starting
- Added 1mL of MagBead buffer to the bead concentrate
- Added 150µl MagBead mix to each sample and pipetted at least 10 times each until completely homogenous
- Placed on shaker for 5 minutes
- Placed on magnet plate, and because it was going slow to the magnet (large volume) the plate was placed on the shaker to help ~5-10 min
- Once the supernatant was clear, the clear liquid was removed from each sample without disturbing the beads while still on the magnet plate
- Still on the magnet, added 200µl of Zymo-Seq Wash buffer to each sample
- Removed 200µl of supernatant from each tube without disturbing the beads while still on the magnet
- Repeated: Still on the magnet, added 200µl of Zymo-Seq Wash buffer to each sample
- Removed 200µl of supernatant from each tube without disturbing the beads while still on the magnet
- Used p20 to remove any remaining liquid in the tubes, and used tips to remove any blobs of wash seen left inside each tube
- Resuspended beads in 10µl DNA elution buffer, and placed in a thermocyler already set at 95°C for 5 minutes
- After incubation, placed tubes back on the magnet plate and removed he ~10µl of supernatant into newly labeled PCR tubes
- Placed in 4 degree fridge overnight as a safe stopping point
P7 Adapter Ligation
- The next day: Thawed reagents L1 and L2 on ice
- Added 10µl of reagent L1 to each tube from yesterday on ice, pipetted to mix, and spun down
- Placed in thermocyler MES login program ZRibo Adapter 1 program:
- 37°C 15 minutes
- 95°C 2 minutes
- 4°C hold
- 95°C 10 min
- 63°C 30 seconds
- 72°C 7 minutes
- 4°C hold
- At hold in program added 20µl reagent L2 to each tube in the thermocycler, pipetted to mix, and spun down
- Pressed enter in the program to continue it
Cleanup with DNA MagBeads
- Added MagBead Buffer to bead concentrate as above
- After program was over, added 60µl MagBeads to each sample and pipetted at least 10 times each until completely homogenous
- Placed on shaker for 5 minutes
- Placed on magnet plate and placed on the shaker to help
- Once the supernatant was clear, the clear liquid was removed from each sample without disturbing the beads while still on the magnet plate
- Still on the magnet, added 150µl of Zymo-Seq Wash buffer to each sample
- Removed 150µl of supernatant from each tube without disturbing the beads while still on the magnet
- Repeated: Still on the magnet, added 150µl of Zymo-Seq Wash buffer to each sample
- Removed 150µl of supernatant from each tube without disturbing the beads while still on the magnet
- Used p20 to remove any remaining liquid in the tubes, and used tips to remove any blobs of wash seen left inside each tube
- Resuspended beads in 10µl DNA Elution Buffer and placed tubes on the shaker for 5 minutes room temp
- After incubation, placed tubes back on the magnet plate and removed he ~10µl of supernatant into newly labeled PCR tubes
P5 Adapter Ligation
- Thawed reagent L3 on ice
- Added 10µl reagent L3 to each sample on ice, pipetted to mix, and spun tubes down
- Placed in thermocyler MES login ZRibo Adapter 2 program:
- 25°C 15 minutes
- 4°C hold
- After program, brought the volume up in each sample tube to 100µl by adding 80µl DNA elution buffer to each tube
Cleanup with DNA MagBeads
- After program was over, added 100µl MagBeads to each sample and pipetted at least 10 times each until completely homogenous
- Placed on shaker for 5 minutes
- Placed on magnet plate and placed on the shaker to help
- Once the supernatant was clear, the clear liquid was removed from each sample without disturbing the beads while still on the magnet plate
- Still on the magnet, added 200µl of Zymo-Seq Wash buffer to each sample
- Removed 200µl of supernatant from each tube without disturbing the beads while still on the magnet
- Repeated: Still on the magnet, added 200µl of Zymo-Seq Wash buffer to each sample
- Removed 200µl of supernatant from each tube without disturbing the beads while still on the magnet
- Used p20 to remove any remaining liquid in the tubes, and used tips to remove any blobs of wash seen left inside each tube
- Resuspended beads in 20µl DNA Elution Buffer and placed tubes on the shaker for 5 minutes room temp
- After incubation, placed tubes back on the magnet plate and removed he ~20µl of supernatant into newly labeled PCR tubes
Library Index PCR
- Thawed 12 index primers included with the kit and the ZymoTaq Premix. Primers have one universal side and one unique side already mixed, at 2.5µM each. Because we got two kits, I used two different primer sets. I equally assigned primers from each set to each treatment because treatments were not randomly distributed through samples but clumped
- Added 5µl of index primers to each sample tube on ice, making sure each tube got a unique primer:
| Sample | Primer Pair |
|---|---|
| 13 | CD Index #7 |
| 14 | CD Index #77 |
| 15 | CD Index #93 |
| 16 | CD Index #5 |
| 17 | CD Index #4 |
| 18 | CD Index #86 |
| 19 | CD Index #12 |
| 20 | CD Index #2 |
| 21 | CD Index #85 |
| 22 | CD Index #94 |
| 23 | CD Index #6 |
| 24 | CD Index #78 |
- Added 25µl ZymoTaq Premix to each sample tube on ice, pipetted to mix, and spun down
- Placed in thermocycler MES login program ZRibo Index Amp program:
- 95°C 10 minutes
- 95°C 30 seconds (13)
- 60°C 30 seconds (13)
- 72°C 1 minute (13)
- 72°C 7 minutes
- 4 hold
Bold steps were cycled through 13 times. I chose 13 because we had previous issues with getting our libraries to amplify enough, and even though I started with more RNA this time we might want to have enough library to save some and send some to sequence. So I increased the cycles by 2 compared to what is recommended for this starting concentration (11)
- After the PCR, increased the volume in each tube to 100µl by add ing 50µl DNA elution buffer
Cleanup with DNA MagBeads
- Added MagBead Buffer to bead concentrate as above
- After program was over, added 685µl MagBeads to each sample and pipetted at least 10 times each until completely homogenous
- Placed on shaker for 5 minutes
- Placed on magnet plate and placed on the shaker to help
- Once the supernatant was clear, the clear liquid was removed from each sample without disturbing the beads while still on the magnet plate
- Still on the magnet, added 200µl of Zymo-Seq Wash buffer to each sample
- Removed 200µl of supernatant from each tube without disturbing the beads while still on the magnet
- Repeated: Still on the magnet, added 200µl of Zymo-Seq Wash buffer to each sample
- Removed 200µl of supernatant from each tube without disturbing the beads while still on the magnet
- Used p20 to remove any remaining liquid in the tubes, and used tips to remove any blobs of wash seen left inside each tube
- Resuspended beads in 22µl DNA Elution Buffer and placed tubes on the shaker for 5 minutes room temp
- After incubation, placed tubes back on the magnet plate and removed the ~22µl of supernatant into newly labeled PCR tubes
QC
D5000 TapeStation following protocol
Full Results
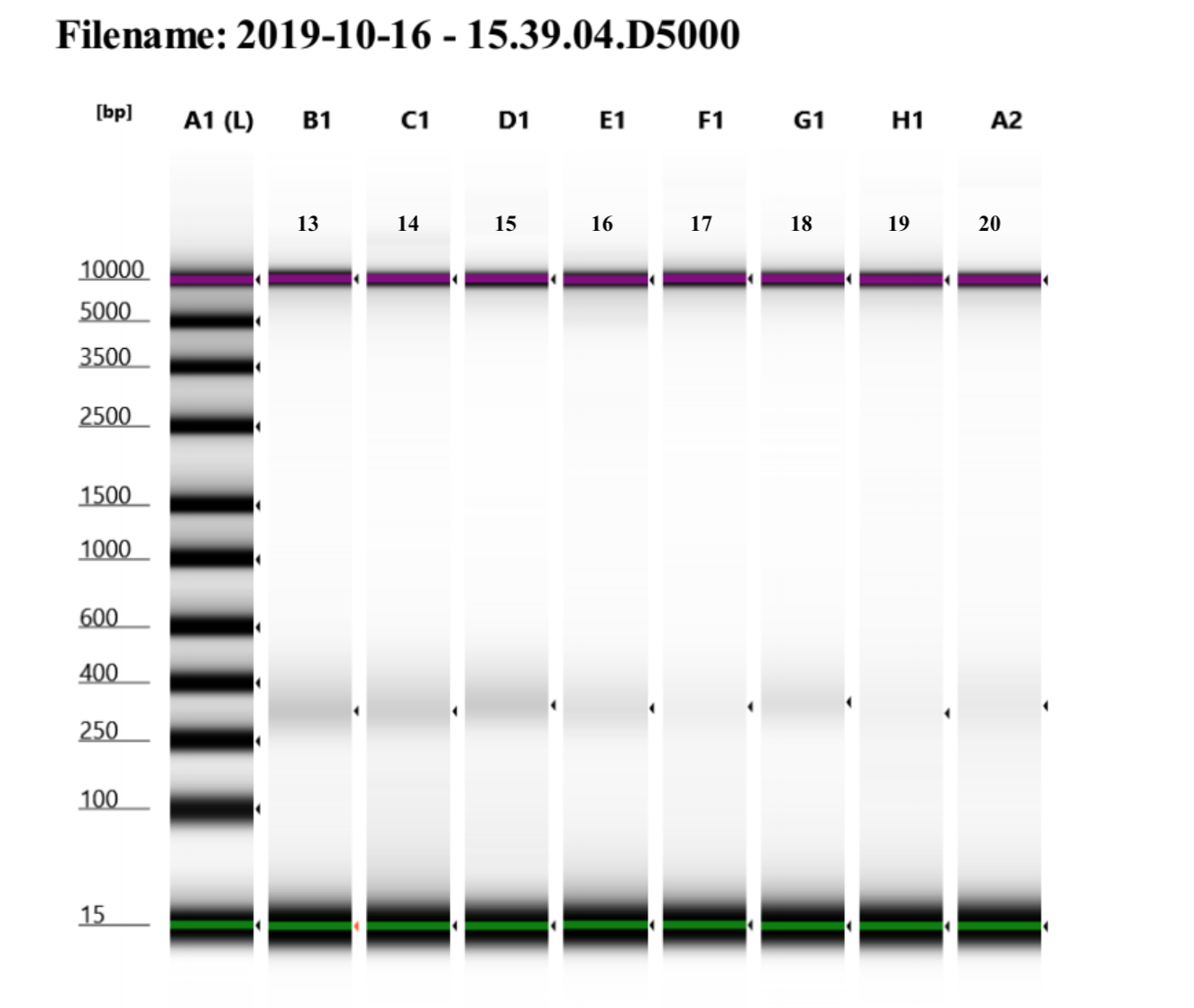
Not all samples were run on a tape because I did not have time to do another tape, but all samples have the same pattern of very low concentrations of libraries but they look to be the right size. I decided to do additional amplifications for each library to try to increase the concentrations.
Additional PCRs
10µl of library was used with 2.5µl of primers and 12.5µl of ZymoTaq Premix for an additional 6 cycle PCR step on all samples. Then using KAPA Pure beads (0.85X) and ethanol washes the samples were cleaned post-PCR and tapestationed again.
Full Results
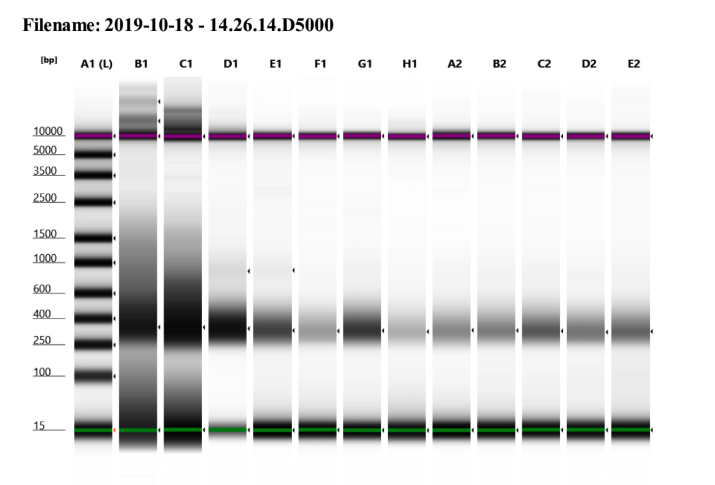
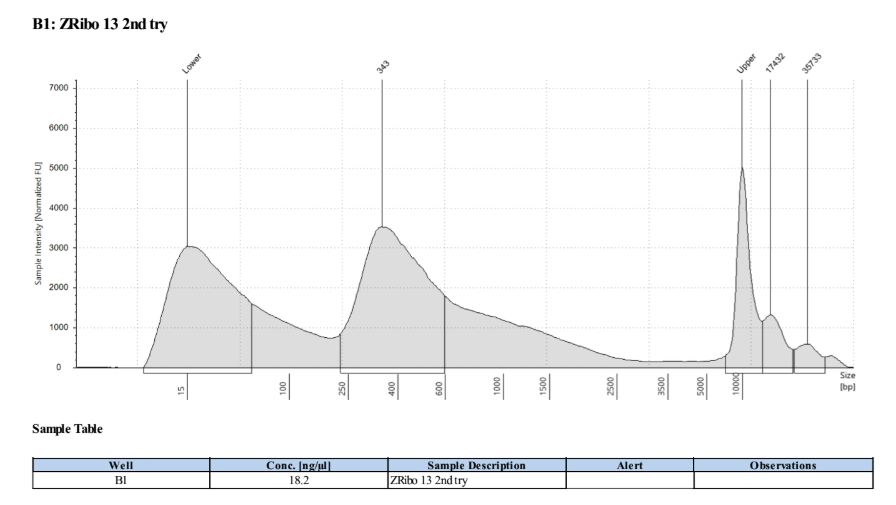
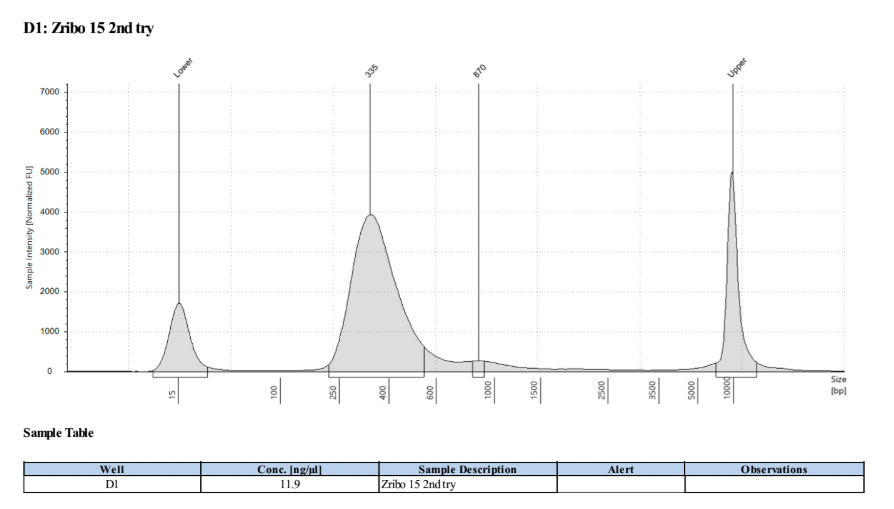
I am not sure why samples 13 and 14 have such weird traces, it may be a tape or buffer error because of how the upper and lower markers spread so much. I will re-tapestation those two samples.
I would say the other samples are good enough to send off for sequencing.
Re-tapestation of samples 13 and 14
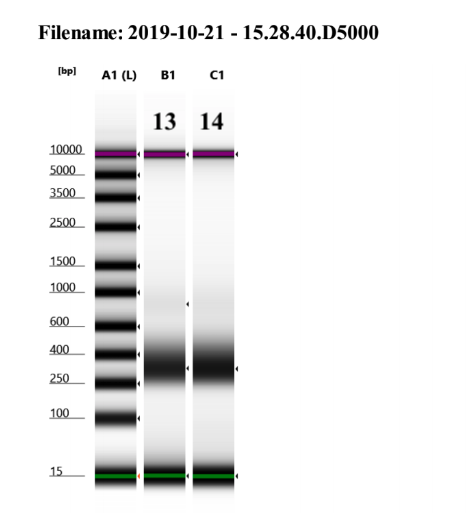
These libraries look like the others!
| ID | ng/ul | volume | total ng |
|---|---|---|---|
| 13 | 8.9 | 19 | 169.1 |
| 14 | 10 | 19 | 190 |
| 15 | 11.7 | 19 | 222.3 |
| 16 | 6.59 | 19 | 125.21 |
| 17 | 2.81 | 19 | 53.39 |
| 18 | 7.36 | 19 | 139.84 |
| 19 | 2.27 | 19 | 43.13 |
| 20 | 3.02 | 19 | 57.38 |
| 21 | 3.68 | 19 | 69.92 |
| 22 | 5.09 | 19 | 96.71 |
| 23 | 4.1 | 19 | 77.9 |
| 24 | 4.54 | 19 | 86.26 |