Primer Resuspention and Test PCR for mtORF Pocillopra Species Determination
Resuspending RORF, Fatp6.1, POCHistoneF and POCHistoneR primers. Testing 5 samples from Mo’orea Connect project with mtORF PCR and gel.
Goal: Resuspend primers to stock and working stock. Try PCR for the mtORF primers (Fatp6.1 and RORF) and modify volumes from paper with PCR protocol that worked for ITS2 amplification.
Major Results: The PCR works, might have over amplification because they cycles were 40
Major Take Aways: I should have done a 100ul reaction then split into 3 tubes like the ITS2 protocol instead of 2 100ul reactions. But amplification was in every sample, probably not a lot of troubleshooting for this, will want to try one more test with 30 cycles and with Danielle’s Pocillopora samples
Primer resuspention and working stocks 2020-07-23
Set Up
- Resuspending with Low TE buffer
- Want to calculate 200uM stock solution
- Centrifuge lofilized oligos for 1 min to collect at the bottom
- Equation is:
ul Low TE = nmol/200 * 1000
Primer Concentration Calculations
- PocHistoneF: 79nmol / 200 * 1000 = 395ul low TE
- PocHistoneR: 91.6nmol / 200 * 1000 = 458ul low TE
- Fatp6.1: 77.3nmol / 200 * 1000 = 386.5ul low TE
-
RORF: 90.4nmol / 200 * 1000 = 452ul low TE
- Added appropriate volume low TE to the tube from IDT
- Vortexed each tube for 15 seconds after TE addition
- Wrote 200uM on the lid
Working Stock Dilution
- Volume wanted: 100ul
- Amount primer wanted: 10uM
- Volume 200uM primer needed: 5ul
-
Volume nuclease-free water: 95ul
- Set up strip tubes with 95ul nuclease free water in 4 of them
- Added 5ul of 200uM primer to labeled tube in strip
- Vortexed and spun down
- Working stock stored in yellow rack in -20
- Resuspended stock stored in Putnam amplicon primer box in -20
Testing Amplifications with Fatp6.1 and RORF 2020-07-26
- Picked 5 samples from Mo’orea connect sequenced samples that have a range of DNA concentrations and sites:
| Sample | Site | DNA conc. |
|---|---|---|
| 38 | 2 | 20.5 |
| 65 | 2 | 56.2 |
| 172 | 4 | 127 |
| 190 | 5 | 76.9 |
| 234 | 3 | 45.9 |
- Planned for a 100ul reaction and to do each sample twice, and do two control samples (no DNA)
- In the paper their reaction volume is 15ul, so I scaled up the volume of primers to fit into a 100ul reaction:
100ul/15ul = X/0.195X = 1.3ul of each primer - But I did not scale up the amount of DNA because they used 1.5ul in their reaction and scaling that up is 6.6ul of DNA, which is a lot more input than there usually in PCRs. I decided to stick with 1.5ul DNA
Master Mix
- 50ul 2X Phusion mix * 12.2 = 610ul
- 1.3ul 10uM FatP6.1 * 12.2 = 15.86ul
- 1.3ul 10uM RORF * 12.2 = 15.86ul
-
45.9ul ultra pure water * 12.2 = 559.98ul
- Set up 2 PCR strip tubes, labeled them with sample numbers and control
- Added 98.5ul of master mix to each labeled tube
- Added 1.5ul DNA of each sample to their tubes
- Added 1.5ul of ultra pure water to their tubes
- Vortexed the strip tubes and spun them down in the minifuge
- Placed in FATP6 RORF program under the Putnam login on the thermomixer
- Program, bold fields are cycled 40 times:
- 60 seconds 94 degrees C
- 30 seconds 94 degrees C
- 30 seconds 53 degrees C
- 75 seconds 72 degrees C
- 5 minutes 72 degrees C
- Program takes 2 hours and 8 minutes to run
- Once it was done placed the tubes in the 4 degree fridge to gel the next day
Agarose Gel
- Followed protocol for making a 1% small gel
- Used 1kb plus generuler as ladder
- Ran gel for 45 minutes at 100volts
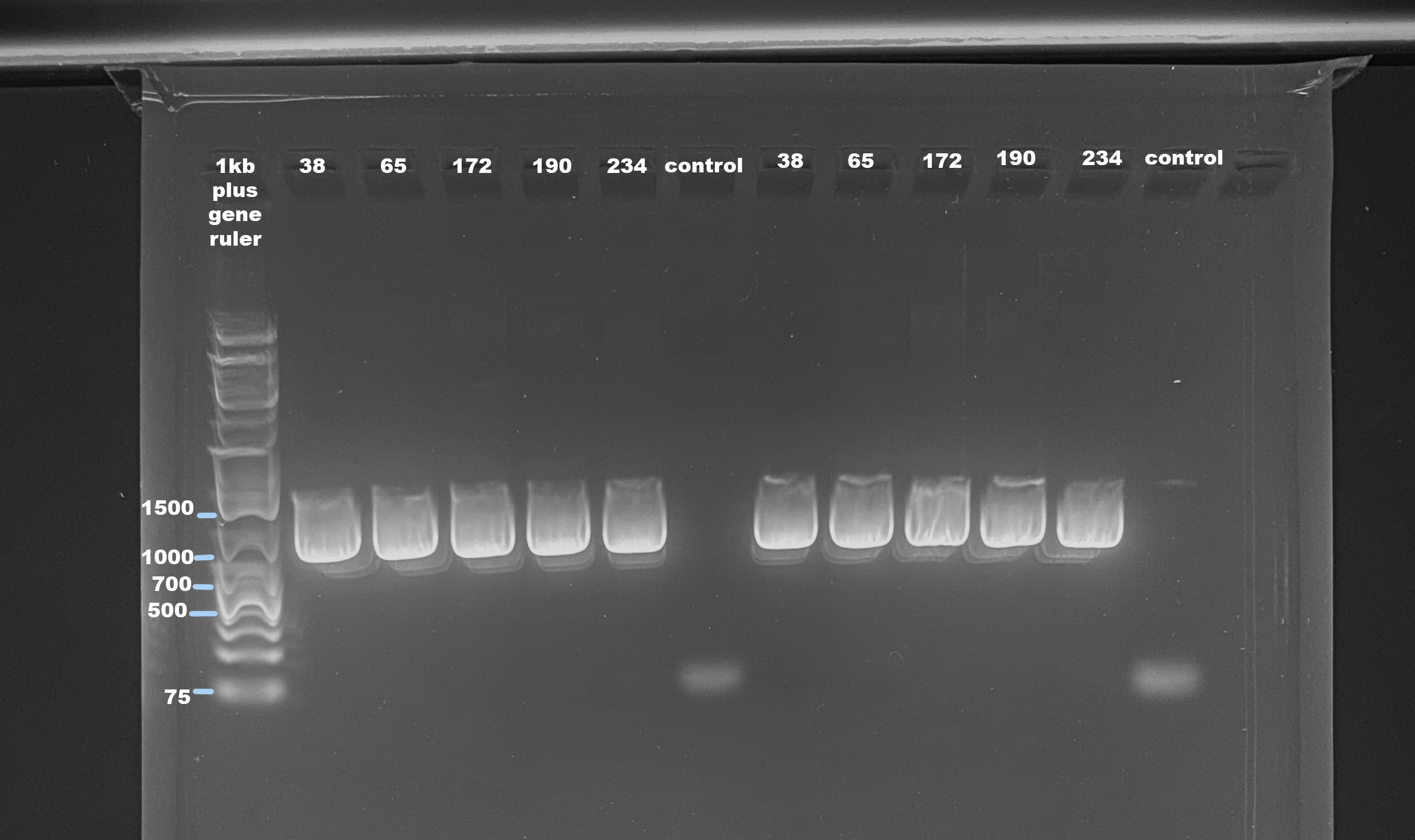
The mtORF should be between 900-1000bp. This looks like the amplification worked, but I am wondering if the stuff above the brightest area (also the right size for mtORF) is a PCR bubble. 40 cycles is a lot and maybe it over amplified. All samples were at a concentration higher than 10ng/ul, which is the concentration in the ITS2 protocol. I will try reducing cycles and input DNA. The control samples had no amplification that small band is likely the primer.