RRBS Library Prep of Samples for Methylation Comparison
Reduced Representation Bisulfite Sequencing Library Prep with the Zymo Research EZ DNA Methylation RRBS Library Prep Test Kit for Methylation Comparison
Samples for methylation comparison are from the Holobiont Integration Project, and were extracted by Emma Strand or myself, see her notebook posts for the extraction specifications: 20190805 and 20180823, 20190718 and 20190903.
MspI Digestion
- Made PCR tubes with 300ng of DNA, 15ng of Spike-in non-mehtylated eColi DNA, and water to 35.5µl
| Sample | Volume DNA | Volume spike | Volume ultra pure water |
|---|---|---|---|
| 1041 | 4.81 | 6 | 24.69 |
| 1471 | 5.47 | 6 | 24.03 |
| 1637 | 3.46 | 6 | 26.04 |
| 1101 | 11.07 | 5 | 18.43 |
| 1548 | 12.99 | 6 | 16.51 |
| 1628 | 17.65 | 6 | 11.85 |
- Made master mix for digestion
- 10X RRBS buffer 4µl * 6.2 = 24.8µl
- MspI 0.5µl * 6.2 = 3.1µl
- Added 4.5µl of digestion master mix to each sample and pipetted to mix and spun down
- Placed in thermocycler RRBS digestion program: 4 hours at 37 degrees C and then a 4 degree hold
Adapter Ligation
- Made master mix for adapter ligation
- 10X RRBS buffer 1µl * 6.2 = 6.2µl
- rATP 0.5µl * 6.2 = 3.1µl
- MspI 1µl * 6.2 = 6.2µl
- T4 DNA ligase 1µl * 6.2 = 6.2µl
- ultra pure water 6µl * 6.2 = 37.2µl
- Pipetted to mix master mix because ligase enzyme
- Added 9.5µl of master mix to each tube
- added 0.5µl RRBS adapters to each tube (adding separately minimizes adapters ligating to themselves)
- Pipetted to mix
- Put in thermocycler program RRBS adapters overnight:
- 21 degrees C 3 hours
- 37 degrees C 1 hour
- 20 degrees C 1 hour
- 37 degrees C 1 hour
- 20 degrees C 1 hour
- 4 degree hold
- The next morning made gap filling master mix
- Taq DNA Polymerase 0.5µl * 6.2 = 3.1µl
- 5-methylcytosine dNTP Mix 1.5µl * 6.2 = 9.3µl
- Added 2µl of gap filling mix to each tube and pipetted to mix
- Placed in theremocyler program 74 degrees C for 30 minutes
Cleanup with DCC
- Made a 1.5mL tube for each sample, added 7:1 ratio DNA binding buffer, so 364µl of DNA binding buffer
- Put elution buffer in thermomixer 56 degrees
- Added DNA sample (52µl) to the appropriate 1.5mL tube
- Vortexed, spun down, and added to the column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 30 seconds, discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged 12,000 rcf 1 minute and 30 seconds, discarded flowthrough
- Transferred columns to 1.5mL tubes
- Added 20µl warmed elution buffer to each column directly
- Incubated 1 minute
- Centrifuged 12,000 rcf 30 seconds
Bisulfite Conversion
- Transferred each sample to PCR tubes
- Added 130µl lightning conversion reagent to each sample in at PCR tube
- Put tubes in the thermocycler Pico bisulfite conversion program (the same for this kit)
Cleanup
- Made 6 spin column, one for each sample
- Added 600µl M-binging buffer each to 6 spin columns
- Added 150µl of the BS reaction (all) to each individual tube
- Invert columns to mix
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 100µl M-Wash buffer to each column
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 200µl L-desulfonation buffer to each column
- Let them sit for 15 minutes
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged columns at 12,000 rcf for 30 seconds and discarded flowthrough
- Added 200µl M-wash buffer to each column
- Centrifuged columns at 12,000 rcf for 1 minute and 30 seconds and discarded flowthrough
- Added 24µl warmed (56C) DNA elution buffer to each column and let sit for 1 minute in new 1.5mL tubes to collect
- Centrifuged columns at 12,000 rcf for 30 seconds
Index Primer Amplification
- Transferred 24µl of DNA to new PCR tubes
- Added 25µl of Library Amp Master mix to each tube
- Added 0.5µl of each planned primer pairs:
| Sample | i5 | i7 |
|---|---|---|
| 1041 | 4 | 4 |
| 1471 | 5 | 5 |
| 1637 | 6 | 6 |
| 1101 | 13 | 13 |
| 1548 | 14 | 14 |
| 1628 | 15 | 15 |
- Placed in theremocyler RRBS index amp program for 11 cycles:
- 94 for 30 sec
- 94 for 30 sec (11)
- 55 for 30 sec (11)
- 68 for 1 min (11)
- 68 for 5 min
- 4 hold
KAPA Pure Bead Cleanup
- Took beads out of fridge beforehand and made fresh 80% EtOH
- Added 50µl (1X) of Kapa Pure beads to each sample
- Performed normal bead cleanup
- Resuspended and eluted samples in 16µl DNA elution buffer
D5000 TapeStation
Full Results
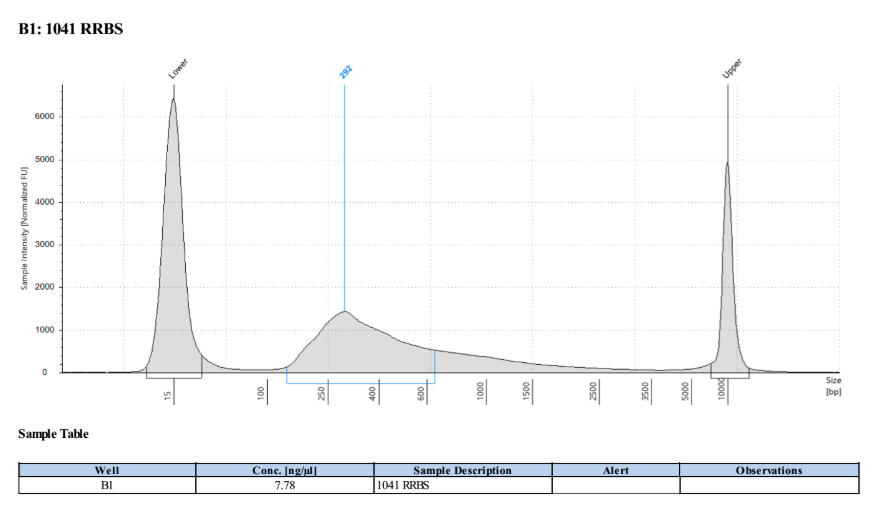
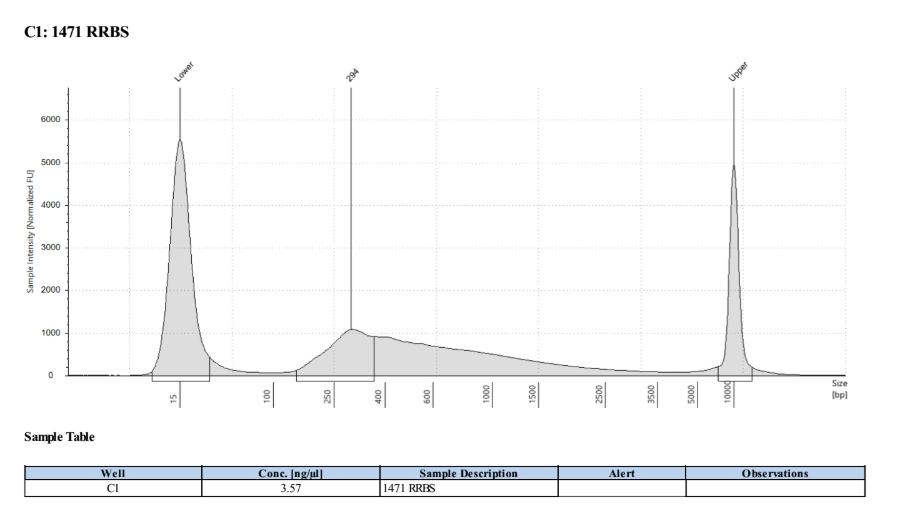
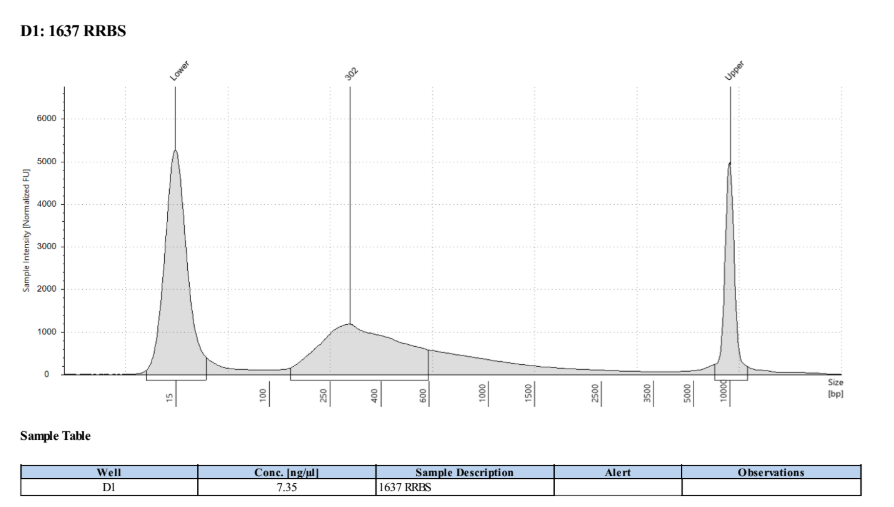
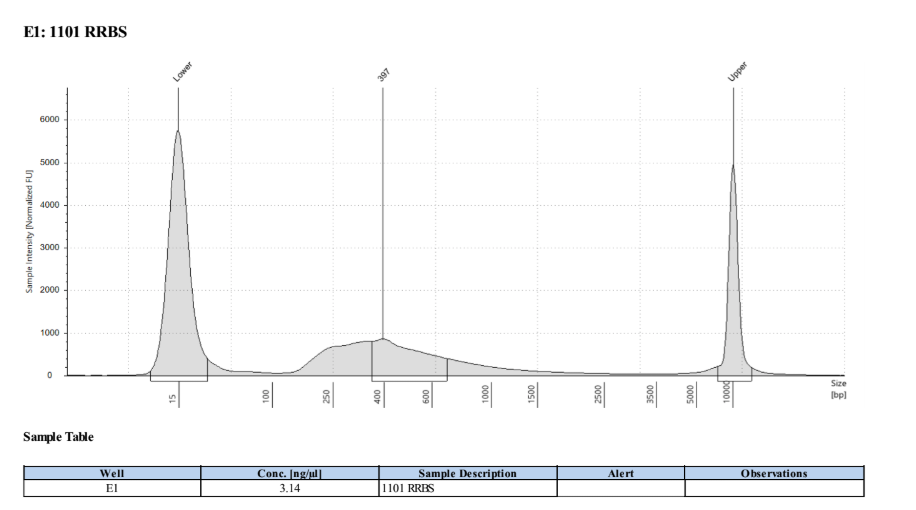
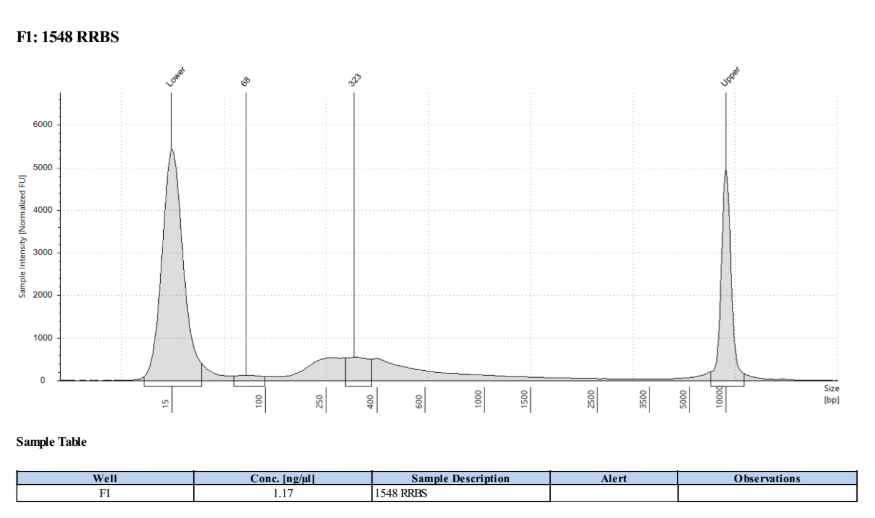
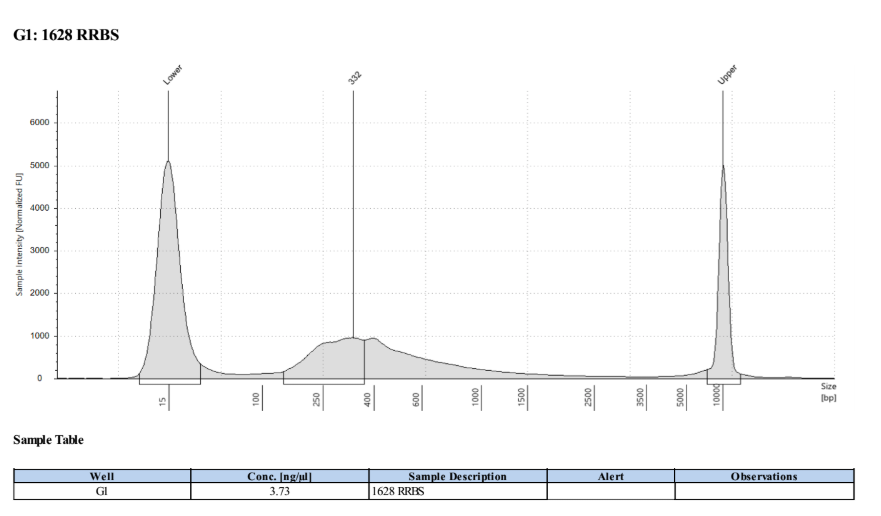
They have long tails but they all look like they worked!!
Written on October 24, 2019