Restriction Enzyme Digest Test for Mo'orea Porites and Pocillopora ddRAD and EpiRAD
12 Tests of Individual and Double Enzymes for Both Species
Two DNA samples of each Pocillopora and Porites were chosen that have high concentrations to use for the tests.
For each sample the following digestions were prepared:
For one sample of each species the following double digestions were prepared:
Master Mixes of the enzymes and buffers were made for each digestion
Master Mix 1 (MM1)
- 10µl Cutsmart buffer * 4.2 = 42µl
- 1µl PstI * 4.2 = 4.2µl
Master Mix 2 (MM2)
- 10µl Cutsmart buffer * 4.2 = 42µl
- 1µl MspI * 4.2 = 4.2µl
Master Mix 3 (MM3)
- 10µl Cutsmart buffer * 2.1 = 21µl
- 1µl PstI * 2.1 = 2.1µl
- 1µl MspI * 2.1 = 2.1µl
Master Mix 4 (MM4)
- 10µl Cutsmart buffer * 2.1 = 21µl
- 1µl PstI * 2.1 = 2.1µl
- 1µl HpaII * 2.1 = 2.1µl
All digestions were done in .2mL strip tubes, and samples were diluted to 500ng in 88µl 10mM Tris HCl to add up to 100µl digestion volume with the addition of the master mix.
| Tube Label | Digestion | Volume Sample (µl) | Volume Tris (µl) | Volume Master Mix |
|---|---|---|---|---|
| SP1 | Pocillopora 172 PstI | 3.94 | 84.06 | 11µl MM1 |
| SP2 | Pocillopora 188 PstI | 4.52 | 83.48 | 11µl MM1 |
| SP3 | Porites 268 PstI | 5.98 | 82.02 | 11µl MM1 |
| SP4 | Porites 199 PstI | 6.93 | 81.07 | 11µl MM1 |
| SM1 | Pocillopora 172 MspI | 3.94 | 84.06 | 11µl MM2 |
| SM2 | Pocillopora 188 MspI | 4.52 | 83.48 | 11µl MM2 |
| SM3 | Porites 268 MspI | 5.98 | 82.02 | 11µl MM2 |
| SM4 | Porites 199 MspI | 6.93 | 81.07 | 11µl MM2 |
| D1 | Pocillopora 172 PstI & MspI | 3.94 | 84.06 | 12µl MM3 |
| D2 | Porites 268 PstI & MspI | 5.98 | 82.02 | 12µl MM3 |
| E1 | Pocillopora 172 PstI & HpaII | 3.94 | 84.06 | 12µl MM4 |
| E2 | Porites 268 PstI & HpaII | 5.98 | 82.02 | 12µl MM4 |
Everything was pipetted repeatedly and gently to mix
Tubes were spun down, and put into a PCR program for 37 degrees C for 12 hours and a hold at 4 degrees C
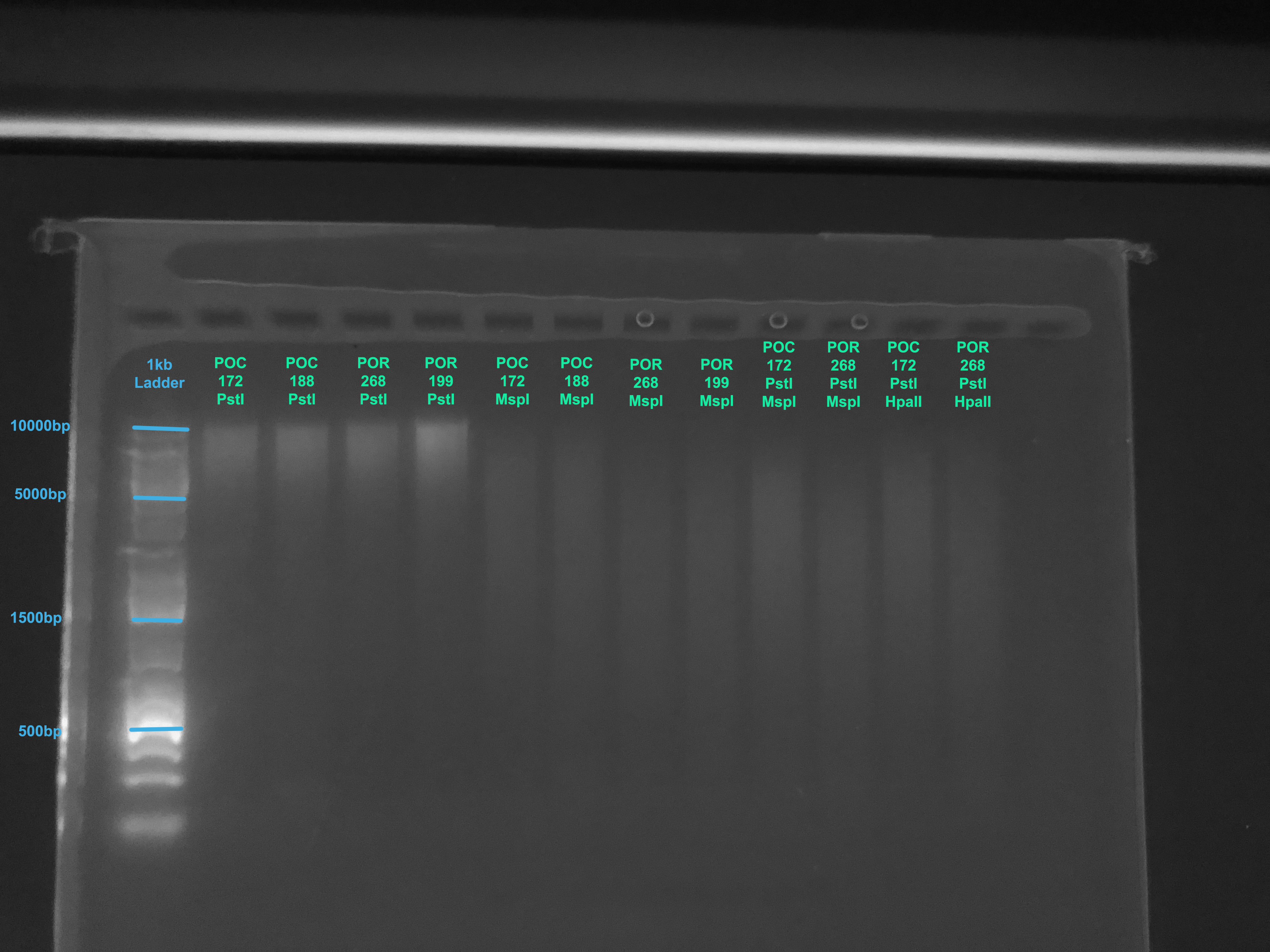
Our TapeStation is down so we used the Bioanalyzer in the GSC to look at the fragment size distributions from the digestions
A High Sensitivity DNA Assay was ran with 1µl of DNA from all of the digestions but SP4 (there are only 11 spots on the chip)
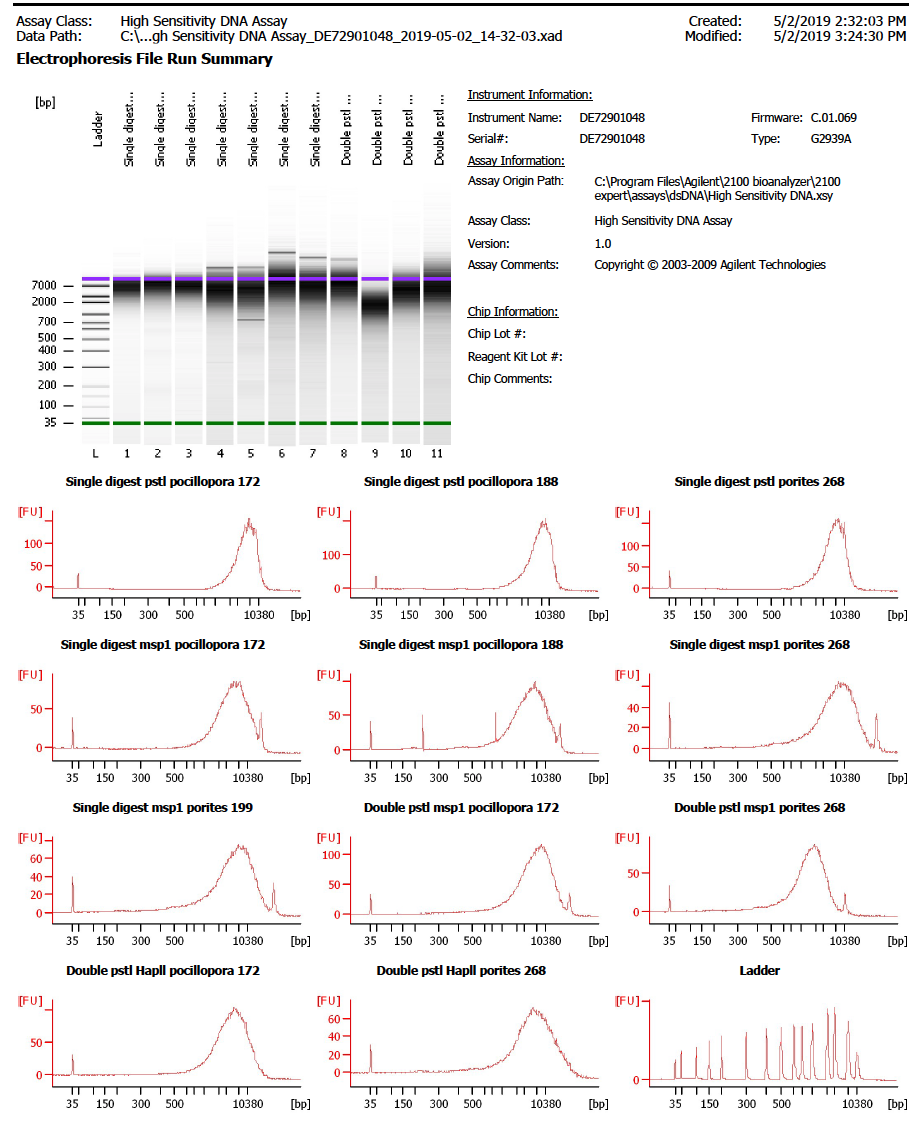
It looks like the upper marker gets off starting at lane 4, so it is hard to tell what the fragment distribution really is. The full pdf of of the Bioanalyzer output is here
A 1.5% gel was also run on the samples to get a better look at them using this protocol